Deck 25: Dna Metabolism
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Match between columns
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/59
Play
Full screen (f)
Deck 25: Dna Metabolism
1
An Okazaki fragment is a:
A)fragment of DNA resulting from endonuclease action.
B)fragment of RNA that is a subunit of the 30S ribosome.
C)piece of DNA that is synthesized in the 3' 5' direction.
D)segment of DNA that is an intermediate in the synthesis of the lagging strand.
E)segment of mRNA synthesized by RNA polymerase.
A)fragment of DNA resulting from endonuclease action.
B)fragment of RNA that is a subunit of the 30S ribosome.
C)piece of DNA that is synthesized in the 3' 5' direction.
D)segment of DNA that is an intermediate in the synthesis of the lagging strand.
E)segment of mRNA synthesized by RNA polymerase.
segment of DNA that is an intermediate in the synthesis of the lagging strand.
2
When a DNA molecule is described as replicating bidirectionally,that means that it has two:
A)chains.
B)independently replicating segment.
C)origins.
D)replication forks.
E)termination points.
A)chains.
B)independently replicating segment.
C)origins.
D)replication forks.
E)termination points.
replication forks.
3
In base-excision repair,the first enzyme to act is:
A)AP endonuclease.
B)Dam methylase.
C)DNA glycosylase.
D)DNA ligase.
E)DNA polymerase.
A)AP endonuclease.
B)Dam methylase.
C)DNA glycosylase.
D)DNA ligase.
E)DNA polymerase.
DNA glycosylase.
4
The role of the Dam methylase is to:
A)add a methyl group to uracil,converting it to thymine.
B)modify the template strand for recognition by repair systems.
C)remove a methyl group from thymine.
D)remove a mismatched nucleotide from the template strand.
E)replace a mismatched nucleotide with the correct one.
A)add a methyl group to uracil,converting it to thymine.
B)modify the template strand for recognition by repair systems.
C)remove a methyl group from thymine.
D)remove a mismatched nucleotide from the template strand.
E)replace a mismatched nucleotide with the correct one.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
5
The function of the eukaryotic DNA replication factor PCNA (proliferating cell nuclear antigen)is similar to that of the -subunit of bacterial DNA polymerase III in that it:
A)facilitates replication of telomeres.
B)forms a circular sliding clamp to increase the processivity of replication.
C)has a 3' 5' proofreading activity.
D)increases the speed but not the processivity of the replication complex.
E)participates in DNA repair.
A)facilitates replication of telomeres.
B)forms a circular sliding clamp to increase the processivity of replication.
C)has a 3' 5' proofreading activity.
D)increases the speed but not the processivity of the replication complex.
E)participates in DNA repair.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
6
Which of the following is not required for initiation of DNA replication in E.coli?
A)DnaB (helicase)
B)DnaG (primase)
C)Dam methylase
D)DNA ligase
E)DnaA (a AAA+ ATPase)
A)DnaB (helicase)
B)DnaG (primase)
C)Dam methylase
D)DNA ligase
E)DnaA (a AAA+ ATPase)

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
7
When bacterial DNA replication introduces a mismatch in a double-stranded DNA,the methyl-directed repair system:
A)cannot distinguish the template strand from the newly replicated strand.
B)changes both the template strand and the newly replicated strand.
C)corrects the DNA strand that is methylated.
D)corrects the mismatch by changing the newly replicated strand.
E)corrects the mismatch by changing the template strand.
A)cannot distinguish the template strand from the newly replicated strand.
B)changes both the template strand and the newly replicated strand.
C)corrects the DNA strand that is methylated.
D)corrects the mismatch by changing the newly replicated strand.
E)corrects the mismatch by changing the template strand.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
8
The proofreading function of DNA polymerase involves all of the following except:
A)a 3' 5' exonuclease.
B)base pairing.
C)detection of mismatched base pairs.
D)phosphodiester bond hydrolysis.
E)reversal of the polymerization reaction.
A)a 3' 5' exonuclease.
B)base pairing.
C)detection of mismatched base pairs.
D)phosphodiester bond hydrolysis.
E)reversal of the polymerization reaction.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
9
At replication forks in E.coli:
A)DNA helicases make endonucleolytic cuts in DNA.
B)DNA primers are degraded by exonucleases.
C)DNA topoisomerases make endonucleolytic cuts in DNA.
D)RNA primers are removed by primase.
E)RNA primers are synthesized by primase.
A)DNA helicases make endonucleolytic cuts in DNA.
B)DNA primers are degraded by exonucleases.
C)DNA topoisomerases make endonucleolytic cuts in DNA.
D)RNA primers are removed by primase.
E)RNA primers are synthesized by primase.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
10
Which of the following is not required for elongation during DNA replication in E.coli?
A)DnaB (helicase)
B)DnaG (primase)
C)DnaC
D)( -sliding clamp)
E)Clamp loader
A)DnaB (helicase)
B)DnaG (primase)
C)DnaC
D)( -sliding clamp)
E)Clamp loader

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
11
Prokaryotic DNA polymerase III:
A)contains a 5' 3' proofreading activity to improve the fidelity of replication.
B)does not require a primer molecule to initiate replication.
C)has a subunit that acts as a circular clamp to improve the processivity of DNA synthesis.
D)synthesizes DNA in the 3' 5' direction.
E)synthesizes only the leading strand;DNA polymerase I synthesizes the lagging strand.
A)contains a 5' 3' proofreading activity to improve the fidelity of replication.
B)does not require a primer molecule to initiate replication.
C)has a subunit that acts as a circular clamp to improve the processivity of DNA synthesis.
D)synthesizes DNA in the 3' 5' direction.
E)synthesizes only the leading strand;DNA polymerase I synthesizes the lagging strand.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
12
The 5' 3' exonuclease activity of E.coli DNA polymerase I is involved in:
A)formation of a nick at the DNA replication origin.
B)formation of Okazaki fragments.
C)proofreading of the replication process.
D)removal of RNA primers by nick translation.
E)sealing of nicks by ligase action.
A)formation of a nick at the DNA replication origin.
B)formation of Okazaki fragments.
C)proofreading of the replication process.
D)removal of RNA primers by nick translation.
E)sealing of nicks by ligase action.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
13
E.coli DNA polymerase III:
A)can initiate replication without a primer.
B)is efficient at nick translation.
C)is the principal DNA polymerase in chromosomal DNA replication.
D)represents over 90% of the DNA polymerase activity in E.coli cells.
E)requires a free 5'-hydroxyl group as a primer.
A)can initiate replication without a primer.
B)is efficient at nick translation.
C)is the principal DNA polymerase in chromosomal DNA replication.
D)represents over 90% of the DNA polymerase activity in E.coli cells.
E)requires a free 5'-hydroxyl group as a primer.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
14
The ABC excinuclease is essential in:
A)base-excision repair.
B)methyl-directed repair.
C)mismatch repair.
D)nucleotide-excision repair.
E)SOS repair.
A)base-excision repair.
B)methyl-directed repair.
C)mismatch repair.
D)nucleotide-excision repair.
E)SOS repair.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
15
In contrast to bacteria,eukaryotic chromosomes need multiple DNA replication origins because:
A)eukaryotic chromosomes cannot usually replicate bidirectionally.
B)eukaryotic genomes are not usually circular,like the bacterial chromosome is.
C)the processivity of the eukaryotic DNA polymerase is much less than the bacterial enzyme.
D)their replication rate is much slower,and it would take too long with only a single origin per chromosome.
E)they have a variety of DNA polymerases for different purposes,and need a corresponding variety of replication origins.
A)eukaryotic chromosomes cannot usually replicate bidirectionally.
B)eukaryotic genomes are not usually circular,like the bacterial chromosome is.
C)the processivity of the eukaryotic DNA polymerase is much less than the bacterial enzyme.
D)their replication rate is much slower,and it would take too long with only a single origin per chromosome.
E)they have a variety of DNA polymerases for different purposes,and need a corresponding variety of replication origins.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
16
In a mammalian cell,DNA repair systems:
A)are extraordinarily efficient energetically.
B)are generally absent,except in egg and sperm cells.
C)can repair deletions,but not mismatches.
D)can repair most types of lesions except those caused by UV light.
E)normally repair more than 99% of the DNA lesions that occur.
A)are extraordinarily efficient energetically.
B)are generally absent,except in egg and sperm cells.
C)can repair deletions,but not mismatches.
D)can repair most types of lesions except those caused by UV light.
E)normally repair more than 99% of the DNA lesions that occur.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
17
Which one of the following statements about enzymes that interact with DNA is true?
A)E)coli DNA polymerase I is unusual in that it possesses only a 5' 3' exonucleolytic activity.
B)Endonucleases degrade circular but not linear DNA molecules.
C)Exonucleases degrade DNA at a free end.
D)Many DNA polymerases have a proofreading 5' 3' exonuclease.
E)Primases synthesize a short stretch of DNA to prime further synthesis.
A)E)coli DNA polymerase I is unusual in that it possesses only a 5' 3' exonucleolytic activity.
B)Endonucleases degrade circular but not linear DNA molecules.
C)Exonucleases degrade DNA at a free end.
D)Many DNA polymerases have a proofreading 5' 3' exonuclease.
E)Primases synthesize a short stretch of DNA to prime further synthesis.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
18
The Ames test is used to:
A)detect bacterial viruses.
B)determine the rate of DNA replication.
C)examine the potency of antibiotics.
D)measure the mutagenic effects of various chemical compounds.
E)quantify the damaging effects of UV light on DNA molecules.
A)detect bacterial viruses.
B)determine the rate of DNA replication.
C)examine the potency of antibiotics.
D)measure the mutagenic effects of various chemical compounds.
E)quantify the damaging effects of UV light on DNA molecules.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
19
The Meselson-Stahl experiment established that:
A)DNA polymerase has a crucial role in DNA synthesis.
B)DNA synthesis in E.coli proceeds by a conservative mechanism.
C)DNA synthesis in E.coli proceeds by a semiconservative mechanism.
D)DNA synthesis requires dATP,dCTP,dGTP,and dTTP.
E)newly synthesized DNA in E.coli has a different base composition than the preexisting DNA.
A)DNA polymerase has a crucial role in DNA synthesis.
B)DNA synthesis in E.coli proceeds by a conservative mechanism.
C)DNA synthesis in E.coli proceeds by a semiconservative mechanism.
D)DNA synthesis requires dATP,dCTP,dGTP,and dTTP.
E)newly synthesized DNA in E.coli has a different base composition than the preexisting DNA.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
20
Which of these enzymes is not directly involved in methyl-directed mismatch repair in E.coli?
A)DNA glycosylase
B)DNA helicase II
C)DNA ligase
D)DNA polymerase III
E)Exonuclease I
A)DNA glycosylase
B)DNA helicase II
C)DNA ligase
D)DNA polymerase III
E)Exonuclease I

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
21
Which of the following is not a feature of site-specific recombination?
A)A specific recombinase enzyme is required.
B)The energy of the phosphodiester bond is preserved in covalent enzyme-DNA linkage.
C)Recombination sites have non-palindromic sequences.
D)Formation of Holliday intermediates is required.
E)Insertions or deletions can result from site-specific recombination.
A)A specific recombinase enzyme is required.
B)The energy of the phosphodiester bond is preserved in covalent enzyme-DNA linkage.
C)Recombination sites have non-palindromic sequences.
D)Formation of Holliday intermediates is required.
E)Insertions or deletions can result from site-specific recombination.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
22
In homologous recombination in E.coli,the protein that moves along a double-stranded DNA,unwinding the strands ahead of it and degrading them,is:
A)chi.
B)DNA ligase.
C)RecA protein.
D)RecBCD enzyme.
E)RuvC protein (resolvase).
A)chi.
B)DNA ligase.
C)RecA protein.
D)RecBCD enzyme.
E)RuvC protein (resolvase).

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
23
Which of the following is not a feature of homologous recombination during meiosis?
A)A double strand break
B)Cleavage of two crossover events
C)Alignment of homologous chromosomes
D)Formation of a single Holliday intermediate
E)Exposed 3' ends invade the intact duplex DNA of the homolog
A)A double strand break
B)Cleavage of two crossover events
C)Alignment of homologous chromosomes
D)Formation of a single Holliday intermediate
E)Exposed 3' ends invade the intact duplex DNA of the homolog

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
24
Which of the following statements is false? In vitro,the strand-exchange reaction:
A)can include formation of a Holliday intermediate.
B)is accompanied by ATP hydrolysis.
C)may involve transient formation of a three- or four-stranded DNA complex.
D)needs RecA protein.
E)requires DNA polymerase.
A)can include formation of a Holliday intermediate.
B)is accompanied by ATP hydrolysis.
C)may involve transient formation of a three- or four-stranded DNA complex.
D)needs RecA protein.
E)requires DNA polymerase.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
25
Which mechanism is used to repair a thymidine dimer in DNA?
A)Mismatch repair
B)Base-excision repair
C)Nucleotide-excision repair
D)Direct repair
E)More than one is used for this type of lesion
A)Mismatch repair
B)Base-excision repair
C)Nucleotide-excision repair
D)Direct repair
E)More than one is used for this type of lesion

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
26
In homologous recombination in E.coli,the protein that assembles into long,helical filaments that coat a region of DNA is:
A)DNA methylase.
B)DNA polymerase.
C)histone.
D)RecA protein.
E)RecBCD enzyme.
A)DNA methylase.
B)DNA polymerase.
C)histone.
D)RecA protein.
E)RecBCD enzyme.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
27
DNA replication in E.coli begins at a site in the DNA called the (a)___________.At the replication fork the (b)___________ strand is synthesized continuously while the (c)_________ strand is synthesized discontinuously.On the strand synthesized discontinuously,the short pieces are called (d)____________ fragments.An RNA primer for each of the fragments is synthesized by an enzyme called (e)__________,and this RNA primer is removed after the fragment is synthesized by the enzyme (f)___________,using its (g)_____________ activity.The nicks left behind in this process are sealed by the enzyme (h)_____________.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
28
All known DNA polymerases catalyze synthesis only in the 5' 3' direction.Nevertheless,during semiconservative DNA replication in the cell,they are able to catalyze the synthesis of both daughter chains,which would appear to require synthesis in the 3' 5' direction.Explain the process that occurs in the cell that allows for synthesis of both daughter chains by DNA polymerase.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
29
Nucleotide polymerization appears to be a thermodynamically balanced reaction (because one phosphodiester bond is broken and one is formed).Nevertheless,the reaction proceeds efficiently both in a test tube and in the cell.Explain.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
30
The repair of cyclobutane pyrimidine dimers by bacterial DNA photolyase involves the cofactor:
A)coenzyme A.
B)coenzyme Q.
C)FADH-.
D)pyridoxal phosphate (PLP).
E)thiamine pyrophosphate (TPP).
A)coenzyme A.
B)coenzyme Q.
C)FADH-.
D)pyridoxal phosphate (PLP).
E)thiamine pyrophosphate (TPP).

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
31
All known DNA polymerases can only elongate a preexisting DNA chain (i.e. ,require a primer)but cannot initiate a new DNA chain.Nevertheless,during semiconservative DNA replication in the cell,entirely new daughter DNA chains are synthesized.Explain the process that occurs in the cell that allows for the synthesis of daughter chains by DNA polymerase.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
32
The DNA below is replicated from left to right.Label the templates for leading strand and lagging strand synthesis.
(5')ACTTCGGATCGTTAAGGCCGCTTTCTGT(3')
(3')TGAAGCCTAGCAATTCCGGCGAAAGACA(5')
(5')ACTTCGGATCGTTAAGGCCGCTTTCTGT(3')
(3')TGAAGCCTAGCAATTCCGGCGAAAGACA(5')

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
33
Briefly describe the biochemical role of the following enzymes in DNA replication in E.coli:
(a)DNA helicase;
(b)primase;
(c)the 3' 5' exonuclease activity of DNA polymerase;
(d)DNA 1igase;
(e)topoisomerases;
(f)the 5' 3' exonuclease activity of DNA polymerase I.
(a)DNA helicase;
(b)primase;
(c)the 3' 5' exonuclease activity of DNA polymerase;
(d)DNA 1igase;
(e)topoisomerases;
(f)the 5' 3' exonuclease activity of DNA polymerase I.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
34
A suitable substrate for DNA polymerase is shown below.Label the primer and template,and indicate which end of each strand must be 3' or 5'. 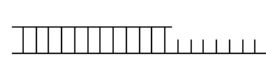 To observe DNA synthesis on this substrate in vitro,what additional reaction components must be added?
To observe DNA synthesis on this substrate in vitro,what additional reaction components must be added?
 To observe DNA synthesis on this substrate in vitro,what additional reaction components must be added?
To observe DNA synthesis on this substrate in vitro,what additional reaction components must be added?
Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
35
An alternative repair system by error-prone translesion DNA synthesis can result in a high mutation rate,because:
A)alternative modified nucleotides can be incorporated more readily.
B)interference from the RecA and SSB proteins hinders the normal replication accuracy.
C)replication proceeds much faster than normal,resulting in many more mistakes.
D)the DNA polymerases involved cannot facilitate base-pairing as well as DNA polymerase III.
E)the DNA polymerases involved lack exonuclease proofreading activities.
A)alternative modified nucleotides can be incorporated more readily.
B)interference from the RecA and SSB proteins hinders the normal replication accuracy.
C)replication proceeds much faster than normal,resulting in many more mistakes.
D)the DNA polymerases involved cannot facilitate base-pairing as well as DNA polymerase III.
E)the DNA polymerases involved lack exonuclease proofreading activities.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
36
What is an Okazaki fragment? What enzyme(s)is (are)required for its formation in E.coli?

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
37
Describe briefly how equilibrium density gradient centrifugation was used to demonstrate that DNA replication in E.coli is semiconservative.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
38
In homologous genetic recombination,RecA protein is involved in:
A)formation of Holliday intermediates and branch migration.
B)introduction of negative supercoils into the recombination products.
C)nicking the two duplex DNA molecules to initiate the reaction.
D)pairing a DNA strand from one duplex DNA molecule with sequences in another duplex,regardless of complementarity.
E)resolution of the Holliday intermediate.
A)formation of Holliday intermediates and branch migration.
B)introduction of negative supercoils into the recombination products.
C)nicking the two duplex DNA molecules to initiate the reaction.
D)pairing a DNA strand from one duplex DNA molecule with sequences in another duplex,regardless of complementarity.
E)resolution of the Holliday intermediate.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
39
Which of the following is false about transposition of DNA?
A)The diversity of immunoglobins is in part due to DNA recombination by transposition.
B)Transposition occurs in both prokaryotes and eukaryotes.
C)Enzymes are not required for transposition.
D)The first step of transposition can be single- or double-stranded DNA cleavage.
E)Transposition can lead to simple movement of a DNA region or duplication of that region in a new location.
A)The diversity of immunoglobins is in part due to DNA recombination by transposition.
B)Transposition occurs in both prokaryotes and eukaryotes.
C)Enzymes are not required for transposition.
D)The first step of transposition can be single- or double-stranded DNA cleavage.
E)Transposition can lead to simple movement of a DNA region or duplication of that region in a new location.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
40
Which mechanism is used to repair a chemically modified base in DNA?
A)Mismatch repair
B)Base-excision repair
C)Nucleotide-excision repair
D)Direct repair
E)More than one is used for this type of lesion
A)Mismatch repair
B)Base-excision repair
C)Nucleotide-excision repair
D)Direct repair
E)More than one is used for this type of lesion

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
41
Why does DNA damage that causes alkylation of nucleotides sometimes lead to transition mutations?

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
42
Outline the four key features of the current model for homologous recombination during meiosis in a eukaryotic cell.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
43
Why is the drug acyclovir effective against the herpes simplex virus?

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
44
Match between columns

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
45
Name the three possible outcomes or consequences (at the DNA level)of a site-specific recombination event.For each of these,explain concisely (in one sentence)how the relative location and orientation of the recombination sites determines the outcome of the recombination event.Do not describe specific examples of site-specific recombination systems.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
46
Outline the key steps that occur during crossing over during meiosis in animal germ-line cells.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
47
What distinguishes the two mechanistic pathways for transposition in bacteria,and what is a cointegrate?

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
48
Explain the role of DNA glycosylases in DNA repair.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
49
Describe the process of nucleotide-excision repair of lesions like pyrimidine dimers in E.coli.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
50
List two proteins or enzymes,other than DNA polymerase III,that are found at the replication fork in E.coli.Describe each of their functions with no more than one sentence.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
51
Briefly explain the difference between base-excision repair and nucleotide-excision repair.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
52
Outline the key steps that occur during meiosis in animal germ-line cells.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
53
The high fidelity of DNA replication is due primarily to immediate error correction by the 3' -> 5' exonuclease (proofreading)activity of the DNA polymerase.Some incorrectly paired bases escape this proofreading,and further errors can arise from challenges to the chemical integrity of the DNA.List the four classes of repair mechanisms that the cell can use to help correct such errors.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
54
Explain how inheriting mutations in genes encoding DNA repair enzymes could lead to increased cancer risk.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
55
What distinguishes the simple from the complex class of bacterial transposon?

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
56
In the bacterial cell,what are catenated chromosomes,when do they arise,and how does the cell resolve the problem posed by their structure?

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
57
List three types of DNA damage that require repair.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
58
DNA synthesis on the lagging strand in E.coli is a complex process known to involve several proteins.Initiation of a new chain is catalyzed by the enzyme (a)_____________,and elongation is catalyzed by the enzyme (b)______________.Synthesis is discontinuous,yielding short segments called (c)_______________,which are eventually joined by the enzyme (d)______________,which requires the cofactor (e)___________.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck
59
Briefly describe the role of recombination in the generation of antibody (immunoglobin)diversity.

Unlock Deck
Unlock for access to all 59 flashcards in this deck.
Unlock Deck
k this deck


