Deck 7: Linkage, Recombination, and Eukaryotic Gene Mapping
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/65
Play
Full screen (f)
Deck 7: Linkage, Recombination, and Eukaryotic Gene Mapping
1
Use the following to answer questions 12-13:
You are doing lab work with a new species of beetle. You have isolated lines that breed true for either blue shells and long antenna or green shells and short antenna. Crossing these lines yields F1 progeny with blue shells and long antenna. Crossing F1 progeny with beetles that have green shells and short antenna yields the following progeny:
Blue shell, long antenna82
Green shell, short antenna78
Blue shell, short antenna37
Green shell, long antenna43
Total240
Assuming that the genes are linked, what is the map distance between them in m.u.?
A) 33.3 m.u.
B) 25.0 m.u.
C) 49.5 m.u.
D) 8.0 m.u.
E) The genes are assorting independently.
You are doing lab work with a new species of beetle. You have isolated lines that breed true for either blue shells and long antenna or green shells and short antenna. Crossing these lines yields F1 progeny with blue shells and long antenna. Crossing F1 progeny with beetles that have green shells and short antenna yields the following progeny:
Blue shell, long antenna82
Green shell, short antenna78
Blue shell, short antenna37
Green shell, long antenna43
Total240
Assuming that the genes are linked, what is the map distance between them in m.u.?
A) 33.3 m.u.
B) 25.0 m.u.
C) 49.5 m.u.
D) 8.0 m.u.
E) The genes are assorting independently.
A
2
Genetic distances within a given linkage group: (Select all that apply.)
A) cannot exceed 100 m.u.
B) are dependent on crossover frequencies between paired, nonsister chromatids.
C) can be measured in centiMorgans or map units.
D) cannot be determined.
E) can only be determined by physical mapping techniques.
A) cannot exceed 100 m.u.
B) are dependent on crossover frequencies between paired, nonsister chromatids.
C) can be measured in centiMorgans or map units.
D) cannot be determined.
E) can only be determined by physical mapping techniques.
B, C
3
Use the following to answer questions 14-15:
Dr. Disney has been raising exotic fruit flies for decades. Recently, he discovered a strain of fruit flies that in the recessive condition have baby blue eyes that he designates as bb. He also has another strain of fruit flies that in the recessive condition have pink wings that are designated as pw. He is able to establish flies that are homozygous for both mutant traits. He mates these two strains with each other. Dr. Disney then takes phenotypically wild-type females from this cross and mates them with double recessive males. In the resulting testcross progeny, he observes 500 flies that are of the following makeup:
41with baby blue eyes and pink wings
207with baby blue eyes only
210with pink wings only
42with wild-type phenotype
Assuming the wild-type alleles for these two genes are b+ and pw+, what is the correct testcross of the F1 flies?
A) b+ pw+/b pw × b pw/b pw
B) b+ pw+/b pw × b pw+/b+ pw
C) b+ pw/b pw+ × b pw/b pw
D) b+ pw/b pw+ × b+ pw+/b pw
E) b+ pw+/b pw × b+ pw/b pw+
Dr. Disney has been raising exotic fruit flies for decades. Recently, he discovered a strain of fruit flies that in the recessive condition have baby blue eyes that he designates as bb. He also has another strain of fruit flies that in the recessive condition have pink wings that are designated as pw. He is able to establish flies that are homozygous for both mutant traits. He mates these two strains with each other. Dr. Disney then takes phenotypically wild-type females from this cross and mates them with double recessive males. In the resulting testcross progeny, he observes 500 flies that are of the following makeup:
41with baby blue eyes and pink wings
207with baby blue eyes only
210with pink wings only
42with wild-type phenotype
Assuming the wild-type alleles for these two genes are b+ and pw+, what is the correct testcross of the F1 flies?
A) b+ pw+/b pw × b pw/b pw
B) b+ pw+/b pw × b pw+/b+ pw
C) b+ pw/b pw+ × b pw/b pw
D) b+ pw/b pw+ × b+ pw+/b pw
E) b+ pw+/b pw × b+ pw/b pw+
C
4
If the recombination frequency between genes (A) and (B) is 5.3%, what is the distance between the genes in map units on the linkage map?
A) 53 m.u.
B) 5.3 m.u.
C) 0.53 m.u.
D) 10.6 m.u.
E) 25 m.u.
A) 53 m.u.
B) 5.3 m.u.
C) 0.53 m.u.
D) 10.6 m.u.
E) 25 m.u.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
5
What major contribution did Barbara McClintock and Harriet Creighton make to the study of recombination?
A) Genetic recombination of alleles is associated with physical exchange between chromosomes.
B) Genes are located on chromosomes and the map distance between them could often be measured by the number of nucleotides in the DNA.
C) Determining map distances in humans could be done by using pedigrees and calculating lod scores.
D) Association studies allow genes that have no obvious phenotype to be accurately mapped.
E) Crossing over does not occur in male Drosophila, so there is no genetic recombination.
A) Genetic recombination of alleles is associated with physical exchange between chromosomes.
B) Genes are located on chromosomes and the map distance between them could often be measured by the number of nucleotides in the DNA.
C) Determining map distances in humans could be done by using pedigrees and calculating lod scores.
D) Association studies allow genes that have no obvious phenotype to be accurately mapped.
E) Crossing over does not occur in male Drosophila, so there is no genetic recombination.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
6
Two linked genes, (A) and (B), are separated by 18 m.u. A man with genotype Aa Bb marries a woman who is aa bb. The man's father was AA BB. What is the probability that their first child will be Aa bb?
A) 0.18
B) 0.41
C) 0.09
D) 0.25
E) 0.50
A) 0.18
B) 0.41
C) 0.09
D) 0.25
E) 0.50

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
7
Linked genes always exhibit:
A) phenotypes that are similar.
B) recombination frequencies of less than 50%.
C) homozygosity when involved in a testcross.
D) a greater number of recombinant offspring than parental offspring when involved in a testcross.
E) a lack of recombinant offspring when a heterozygous parent is involved in a testcross.
A) phenotypes that are similar.
B) recombination frequencies of less than 50%.
C) homozygosity when involved in a testcross.
D) a greater number of recombinant offspring than parental offspring when involved in a testcross.
E) a lack of recombinant offspring when a heterozygous parent is involved in a testcross.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
8
Assume that an individual of AB/ab genotype is involved in a testcross and four classes of testcross progeny are found in equal frequencies. Which of the following statements is TRUE?
A) The genes A and B are on the same chromosome and closely linked.
B) The genes A and B are on the same chromosome and very far apart.
C) The genes A and B are probably between 10 and 20 map units apart on the same chromosome.
D) The genes A and B are likely located on different chromosomes.
E) The genes A and B could be located on different chromosomes or on the same chromosome and very far apart.
A) The genes A and B are on the same chromosome and closely linked.
B) The genes A and B are on the same chromosome and very far apart.
C) The genes A and B are probably between 10 and 20 map units apart on the same chromosome.
D) The genes A and B are likely located on different chromosomes.
E) The genes A and B could be located on different chromosomes or on the same chromosome and very far apart.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
9
Linked genes:
A) assort randomly.
B) cannot cross over and recombine.
C) are allelic.
D) co-segregate.
E) will segregate independently.
A) assort randomly.
B) cannot cross over and recombine.
C) are allelic.
D) co-segregate.
E) will segregate independently.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
10
You are studying two linked genes in lizards. You have two females and you know that both are the same genotype, heterozygous for both genes (A/a and B/b). You testcross each female to a male that is fully homozygous recessive for both genes (a/a and b/b) and get the following progeny with the following phenotypes: How can you explain the drastic difference between these two crosses?
A) The two genes are assorting independently in female 1 and are linked in female 2.
B) The two genes are linked in female 1 and are assorting independently in female 2.
C) The two alleles are in the coupling configuration in female 1 but in the repulsion configuration in female 2.
D) The two alleles are in the repulsion configuration in female 1 but in the coupling configuration in female 2.
E) The two genes are likely located on a sex chromosome in female 1 and are likely located on an autosome in female 2.
A) The two genes are assorting independently in female 1 and are linked in female 2.
B) The two genes are linked in female 1 and are assorting independently in female 2.
C) The two alleles are in the coupling configuration in female 1 but in the repulsion configuration in female 2.
D) The two alleles are in the repulsion configuration in female 1 but in the coupling configuration in female 2.
E) The two genes are likely located on a sex chromosome in female 1 and are likely located on an autosome in female 2.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
11
Use the following to answer questions 12-13:
You are doing lab work with a new species of beetle. You have isolated lines that breed true for either blue shells and long antenna or green shells and short antenna. Crossing these lines yields F1 progeny with blue shells and long antenna. Crossing F1 progeny with beetles that have green shells and short antenna yields the following progeny:
Blue shell, long antenna82
Green shell, short antenna78
Blue shell, short antenna37
Green shell, long antenna43
Total240
A chi-square test is done to test for independent assortment. What is the resulting chi-square value and how many degree(s) of freedom should be used in its interpretation?
A) 27.1 and one degree of freedom
B) 14.9 and three degrees of freedom
C) 14.9 and two degrees of freedom
D) 27.1 and three degrees of freedom
E) 0.42 and two degrees of freedom
You are doing lab work with a new species of beetle. You have isolated lines that breed true for either blue shells and long antenna or green shells and short antenna. Crossing these lines yields F1 progeny with blue shells and long antenna. Crossing F1 progeny with beetles that have green shells and short antenna yields the following progeny:
Blue shell, long antenna82
Green shell, short antenna78
Blue shell, short antenna37
Green shell, long antenna43
Total240
A chi-square test is done to test for independent assortment. What is the resulting chi-square value and how many degree(s) of freedom should be used in its interpretation?
A) 27.1 and one degree of freedom
B) 14.9 and three degrees of freedom
C) 14.9 and two degrees of freedom
D) 27.1 and three degrees of freedom
E) 0.42 and two degrees of freedom

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
12
Assume that A and B are two linked genes on an autosome in Drosophila. A testcross is made where AB/ab flies are crossed to ab/ab flies and the progeny are counted and shown below. However, it is known that the Aa/bb genotype is lethal before the flies hatch and does not appear among the testcross progeny counted. What is the most precise map distance between the two genes that can be calculated from these data? Aa Bb = 235
Aa bb = 225
Aa Bb = 20
A) 4.2 m.u.
B) 4.0 m.u.
C) 16.4 m.u.
D) 8.0 m.u.
E) 50 m.u.
Aa bb = 225
Aa Bb = 20
A) 4.2 m.u.
B) 4.0 m.u.
C) 16.4 m.u.
D) 8.0 m.u.
E) 50 m.u.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
13
What is the relationship with respect to location between the two genes?
A) They are far apart on the same chromosome and assorting independently.
B) They are linked and the map distance between them is 41.5 m.u.
C) They are on different chromosomes and assorting independently.
D) They are linked and 16.6 m.u. apart.
E) They are linked and 50.0 m.u. apart.
A) They are far apart on the same chromosome and assorting independently.
B) They are linked and the map distance between them is 41.5 m.u.
C) They are on different chromosomes and assorting independently.
D) They are linked and 16.6 m.u. apart.
E) They are linked and 50.0 m.u. apart.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
14
Is it possible for two different genes located on the same chromosome to assort independently?
A) No, if two genes are on the same chromosome, they will be linked and the recombination frequency will be less than 50%.
B) Yes, if the two genes are close enough to each other, there will be a limited number of crossover events between them.
C) No, there will be very high crossover interference such that the recombination frequency will be reduced significantly.
D) Yes, if the genes are far enough apart on the same chromosome, a crossover will occur between them in just about every meiotic event.
E) Yes, but only if the two genes are both homozygous.
A) No, if two genes are on the same chromosome, they will be linked and the recombination frequency will be less than 50%.
B) Yes, if the two genes are close enough to each other, there will be a limited number of crossover events between them.
C) No, there will be very high crossover interference such that the recombination frequency will be reduced significantly.
D) Yes, if the genes are far enough apart on the same chromosome, a crossover will occur between them in just about every meiotic event.
E) Yes, but only if the two genes are both homozygous.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
15
A genetic map shows which of the following?
A) the distance in numbers of nucleotides between two genes
B) the number of genes on each of the chromosomes of a species
C) the linear order of genes on a chromosome
D) the location of chromosomes in the nucleus when they line up at metaphase during mitosis
E) the location of double crossovers that occur between two genes
A) the distance in numbers of nucleotides between two genes
B) the number of genes on each of the chromosomes of a species
C) the linear order of genes on a chromosome
D) the location of chromosomes in the nucleus when they line up at metaphase during mitosis
E) the location of double crossovers that occur between two genes

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
16
Recombination frequencies can be calculated by:
A) counting the number of recombinant and parental offspring when an individual who is heterozygous for two genes is involved in a testcross.
B) performing a chi-square test on the F2 progeny when an individual who is homozygous for two genes is initially crossed to an individual who is homozygous recessive for these two genes.
C) counting the number of offspring who are expressing the dominant phenotype when a heterozygous individual for two genes is involved in a testcross.
D) performing a chi-square test of the progeny of a cross between parents who are both heterozygous for the same two genes.
E) counting the number of offspring that are found in the cross of an individual who is heterozygous for two genes with another parent who is homozygous dominant for one of these genes and homozygous recessive for the other gene.
A) counting the number of recombinant and parental offspring when an individual who is heterozygous for two genes is involved in a testcross.
B) performing a chi-square test on the F2 progeny when an individual who is homozygous for two genes is initially crossed to an individual who is homozygous recessive for these two genes.
C) counting the number of offspring who are expressing the dominant phenotype when a heterozygous individual for two genes is involved in a testcross.
D) performing a chi-square test of the progeny of a cross between parents who are both heterozygous for the same two genes.
E) counting the number of offspring that are found in the cross of an individual who is heterozygous for two genes with another parent who is homozygous dominant for one of these genes and homozygous recessive for the other gene.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
17
Crossing over occurs during:
A) late anaphase.
B) prophase.
C) metaphase.
D) early anaphase.
E) telophase.
A) late anaphase.
B) prophase.
C) metaphase.
D) early anaphase.
E) telophase.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
18
Two linked genes, (A) and (B), are separated by 18 m.u. A man with genotype Aa Bb marries a woman who is aa bb. The man's father was AA BB. What is the probability that their first two children will both be ab/ab?
A) 0.168
B) 0.0081
C) 0.032
D) 0.062
E) 0.13
A) 0.168
B) 0.0081
C) 0.032
D) 0.062
E) 0.13

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
19
A testcross includes:
A) one parent who is homozygous recessive for one gene pair and a second parent who is homozygous recessive for a second gene pair.
B) one parent who is homozygous dominant for one or more genes and a second parent who is homozygous recessive for these same genes.
C) two parents who are both heterozygous for two or more genes.
D) one parent who shows the dominant phenotype for one or more genes and a second parent who is homozygous recessive for these genes.
E) one parent who shows the recessive phenotype for one or more genes and a second parent who is homozygous dominant for these genes.
A) one parent who is homozygous recessive for one gene pair and a second parent who is homozygous recessive for a second gene pair.
B) one parent who is homozygous dominant for one or more genes and a second parent who is homozygous recessive for these same genes.
C) two parents who are both heterozygous for two or more genes.
D) one parent who shows the dominant phenotype for one or more genes and a second parent who is homozygous recessive for these genes.
E) one parent who shows the recessive phenotype for one or more genes and a second parent who is homozygous dominant for these genes.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
20
Recombination occurs through:
A) crossing over and chromosome interference.
B) chromosome interference and independent assortment.
C) somatic-cell hybridization and chromosome interference.
D) complete linkage and chromosome interference.
E) crossing over and independent assortment.
A) crossing over and chromosome interference.
B) chromosome interference and independent assortment.
C) somatic-cell hybridization and chromosome interference.
D) complete linkage and chromosome interference.
E) crossing over and independent assortment.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
21
Geneticists often assume that map distances less than 7 to 8 map units (m.u.) are quite accurate. Map distances that exceed this threshold significantly are assumed to be less accurate and the level of accuracy declines as map distances increase. Briefly explain this observation.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
22
Lod scores measure:
A) the relatedness of two individuals.
B) the number of crossover events that occur along an entire chromosome.
C) how often double crossovers occur.
D) the length of a linkage group.
E) the likelihood of linkage between genes.
A) the relatedness of two individuals.
B) the number of crossover events that occur along an entire chromosome.
C) how often double crossovers occur.
D) the length of a linkage group.
E) the likelihood of linkage between genes.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
23
In flower beetles, pygmy (py) is recessive to normal size (py+), and red color (r) is recessive to brown (r+). A beetle heterozygous for these characteristics was test-crossed to a beetle homozygous for pygmy and red. The following are progeny phenotypes from this testcross. Carry out a series of chi-square tests to determine if there is equal segregation of alleles at the py locus.
- What is the correct chi-square value and how many degree(s) of freedom should be used in its interpretation?
A) 0.16 with one degree of freedom
B) 0.16 with three degrees of freedom
C) 0.48 with one degree of freedom
D) 0.48 with two degrees of freedom
E) 4.56 with one degree of freedom
- What is the correct chi-square value and how many degree(s) of freedom should be used in its interpretation?
A) 0.16 with one degree of freedom
B) 0.16 with three degrees of freedom
C) 0.48 with one degree of freedom
D) 0.48 with two degrees of freedom
E) 4.56 with one degree of freedom

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
24
A double heterozygote for two linked genes in the mouse is test-crossed by crossing it to a homozygous recessive parent. In the offspring, the two parental classes appear in a frequency of 45% each, and the two recombinant classes appear in a frequency of 5% each. What is the distance in map units between the two genes?
A) 45 m.u.
B) 22.5 m.u.
C) 10 m.u.
D) 5 m.u.
E) 2.5 m.u.
A) 45 m.u.
B) 22.5 m.u.
C) 10 m.u.
D) 5 m.u.
E) 2.5 m.u.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
25
In flower beetles, pygmy (py) is recessive to normal size (py+), and red color (r) is recessive to brown (r+). A beetle heterozygous for these characteristics was test-crossed to a beetle homozygous for pygmy and red. The following are progeny phenotypes from this testcross. Carry out a series of chi-square tests to determine if the two loci are assorting independently.
-What is the correct chi-square value and how many degree(s) of freedom should be used in its interpretation?
A) 112 with one degree of freedom
B) 265 with three degrees of freedom
C) 367 with four degree of freedom
D) 16.5 with three degrees of freedom
E) 367 with three degrees of freedom
-What is the correct chi-square value and how many degree(s) of freedom should be used in its interpretation?
A) 112 with one degree of freedom
B) 265 with three degrees of freedom
C) 367 with four degree of freedom
D) 16.5 with three degrees of freedom
E) 367 with three degrees of freedom

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
26
Discuss the differences and at least one similarity between recombination and independent assortment.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
27
What is the chi-square test used for, and what does it tell you?

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
28
Two genes, A and B, are located 30 map units apart. The dihybrid shown below is mated to a tester aa bb. What proportion of the offspring is expected to be dominant for both traits?
A) 0%
B) 15%
C) 30%
D) 35%
E) 70%
A) 0%
B) 15%
C) 30%
D) 35%
E) 70%

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
29
In soreflies (a hypothetical insect), the dominant allele, L, is responsible for resistance to a common insecticide called Loritol. Another dominant allele, M, is responsible for the ability of soreflies to sing like birds. A true-breeding mute, Loritol-resistant sorefly, was mated to a true-breeding singing, Loritol-sensitive sorefly, and the singing, Loritol-resistant female progeny were testcrossed with true-breeding wild-type (i.e., mute, Loritol-sensitive) males. Of the 400 total progeny produced, 117 were mute and Loritol-resistant, 114 could sing and were Loritol-sensitive, 83 could sing and were Loritol-resistant, and 86 were mute and Loritol-sensitive. A chi-square test is done to determine if there is equal segregation of alleles at the L locus. What will be the chi-square value obtained and how many degrees of freedom would be used to interpret this value?
A) 0.09 and one degree of freedom
B) 0.56 and two degrees of freedom
C) 0 and one degree of freedom
D) 9.72 and four degrees of freedom
E) A chi-square test is not the appropriate statistical test to answer this question.
A) 0.09 and one degree of freedom
B) 0.56 and two degrees of freedom
C) 0 and one degree of freedom
D) 9.72 and four degrees of freedom
E) A chi-square test is not the appropriate statistical test to answer this question.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
30
A series of two-point crosses among fruit flies is carried out between genes for brown eyes (bw), arc wings (a), vestigial wings (vg), ebony body color (e), and curved wings (cv). The following number of nonrecombinant and recombinant progeny were obtained from each cross: Using these data from two-point crosses, what it the best genetic map (in m.u.) that can be developed?
A) cv 5 bw 13 a 34 vg with e assorting independently
B) bw 5 cv 24 vg 32 a with e assorting independently
C) a 5 bw 13 vg 24 e with vg assorting independently
D) cv 13 bw 5 a 27 vg with e assorting independently
E) bw 5 a 24 cv 13 vg with e assorting independently
A) cv 5 bw 13 a 34 vg with e assorting independently
B) bw 5 cv 24 vg 32 a with e assorting independently
C) a 5 bw 13 vg 24 e with vg assorting independently
D) cv 13 bw 5 a 27 vg with e assorting independently
E) bw 5 a 24 cv 13 vg with e assorting independently

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
31
You are examining the following human pedigree and want to determine if the rare dominant disease allele (D) is linked to a specific DNA sequence location you are using as a molecular marker. Parental and progeny genotypes and phenotypes are indicated. Note that the father is a dihybrid at both loci, but the mother is homozygous at both loci. There is complete penetrance of the trait and a linkage phase of D-R1/d-R2 in the father. Assuming that the marker and the gene are linked, what is the best estimate of the map distance between the two loci? 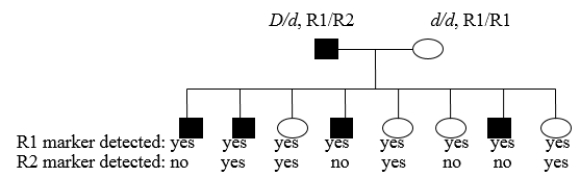
A) 12 m.u.
B) 50 m.u.
C) 16 m.u.
D) 5 m.u.
E) 25 m.u.

A) 12 m.u.
B) 50 m.u.
C) 16 m.u.
D) 5 m.u.
E) 25 m.u.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
32
Assume that you are able to examine a total of 100 separate meiotic events in an animal species. You note that within 10 of these meiotic events, there was a crossover event occurring between genes A and B. In the remaining 90 events there was no crossover event between these two genes. What would be the expected map distance between genes A and B?
A) 10 m.u.
B) 5 m.u.
C) 20 m.u.
D) 45 m.u.
E) 50 m.u.
A) 10 m.u.
B) 5 m.u.
C) 20 m.u.
D) 45 m.u.
E) 50 m.u.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
33
An individual has the following genotype. Gene loci (A) and (B) are 15 m.u. apart. What are the correct frequencies of some of the gametes that can be made by this individual? 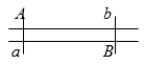
A) Ab = 7.5%; AB = 42.5%
B) ab = 25%; aB = 50%
C) AB = 7.5%; aB = 42.5%
D) aB = 15%; Ab = 70%
E) aB = 70%; Ab = 15%

A) Ab = 7.5%; AB = 42.5%
B) ab = 25%; aB = 50%
C) AB = 7.5%; aB = 42.5%
D) aB = 15%; Ab = 70%
E) aB = 70%; Ab = 15%

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
34
Use the following to answer questions :
You just bought two black guinea pigs, one male and one female, of the same genotype from the pet store that are known to be heterozygous (Bb). You also know that black fur (BB) is dominant over white fur (bb) and that a lethal recessive allele is located only 1 m.u. away from the recessive b allele, and your animals are both heterozygous for this gene also.
You decide to start raising your own guinea pigs, but after mating these animals several times, you discover they produce only black offspring among the first 12 progeny. How would you best explain this result?
A) The B locus is on the X chromosome, so it can never produce a white phenotype.
B) The B allele is actually codominant with the b allele, so a white phenotype cannot be produced.
C) The recessive l allele is in tight coupling linkage with the b allele, so almost all of the bb offspring will also be ll and thus die before they can be observed.
D) The dominant L allele is in tight repulsion linkage with the B allele, so it will be impossible to produce the Bb genotype that would express the white phenotype.
E) Normally, it would be expected that 25% of the offspring would be white, but in this case, random deviations from the expected resulted in no white offspring.
You just bought two black guinea pigs, one male and one female, of the same genotype from the pet store that are known to be heterozygous (Bb). You also know that black fur (BB) is dominant over white fur (bb) and that a lethal recessive allele is located only 1 m.u. away from the recessive b allele, and your animals are both heterozygous for this gene also.
You decide to start raising your own guinea pigs, but after mating these animals several times, you discover they produce only black offspring among the first 12 progeny. How would you best explain this result?
A) The B locus is on the X chromosome, so it can never produce a white phenotype.
B) The B allele is actually codominant with the b allele, so a white phenotype cannot be produced.
C) The recessive l allele is in tight coupling linkage with the b allele, so almost all of the bb offspring will also be ll and thus die before they can be observed.
D) The dominant L allele is in tight repulsion linkage with the B allele, so it will be impossible to produce the Bb genotype that would express the white phenotype.
E) Normally, it would be expected that 25% of the offspring would be white, but in this case, random deviations from the expected resulted in no white offspring.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
35
In soreflies (a hypothetical insect), the dominant allele, L, is responsible for resistance to a common insecticide called Loritol. Another dominant allele, M, is responsible for the ability of soreflies to sing like birds. A true-breeding mute, Loritol-resistant sorefly was mated to a true-breeding singing, Loritol-sensitive sorefly, and the singing, Loritol-resistant female progeny were test-crossed with true-breeding wild-type (i.e., mute, Loritol-sensitive) males. Of the 400 total progeny produced, 117 were mute and Loritol-resistant, 114 could sing and were Loritol-sensitive, 83 could sing and were Loritol-resistant, and 86 were mute and Loritol-sensitive. What will be the results of a chi-square test for independent assortment?
A) 9.70 with three degrees of freedom
B) 4.63 with three degrees of freedom
C) 6.48 with four degrees of freedom
D) 2.54 with one degree of freedom
E) Because there are four classes of offspring, the genes must be assorting independently.
A) 9.70 with three degrees of freedom
B) 4.63 with three degrees of freedom
C) 6.48 with four degrees of freedom
D) 2.54 with one degree of freedom
E) Because there are four classes of offspring, the genes must be assorting independently.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
36
Use the following to answer questions :
You just bought two black guinea pigs, one male and one female, of the same genotype from the pet store that are known to be heterozygous (Bb). You also know that black fur (BB) is dominant over white fur (bb) and that a lethal recessive allele is located only 1 m.u. away from the recessive b allele, and your animals are both heterozygous for this gene also.
What is the probability of finding a white individual among the progeny if you cross these two animals?
A) 0.25
B) 0.0025
C) 0.000025
D) 0.005
E) 0.495
You just bought two black guinea pigs, one male and one female, of the same genotype from the pet store that are known to be heterozygous (Bb). You also know that black fur (BB) is dominant over white fur (bb) and that a lethal recessive allele is located only 1 m.u. away from the recessive b allele, and your animals are both heterozygous for this gene also.
What is the probability of finding a white individual among the progeny if you cross these two animals?
A) 0.25
B) 0.0025
C) 0.000025
D) 0.005
E) 0.495

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
37
In corn, small pollen (sp) is recessive to normal pollen (sp+) and banded necrotic tissue, called zebra necrotic (zn), is recessive to normal tissue (zn+). The genes that produce these phenotypes are closely linked on chromosome 10. If no crossing over occurs between these two loci, give the types of progeny expected from the following cross:
A) sp+ zn+/sp zn; sp zn/sp zn
B) sp+ zn/sp zn; sp zn+/sp zn
C) sp+ zn+/sp+ zn+; sp+ zn+/sp zn; sp zn/sp zn
D) sp+ zn/sp+ zn; sp+ zn/sp zn+; sp zn+/sp+ zn; sp zn+/sp zn+
E) sp+ zn+/sp zn; sp+ zn/sp zn
A) sp+ zn+/sp zn; sp zn/sp zn
B) sp+ zn/sp zn; sp zn+/sp zn
C) sp+ zn+/sp+ zn+; sp+ zn+/sp zn; sp zn/sp zn
D) sp+ zn/sp+ zn; sp+ zn/sp zn+; sp zn+/sp+ zn; sp zn+/sp zn+
E) sp+ zn+/sp zn; sp+ zn/sp zn

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
38
What does lod stand for?
A) linkage over DNA
B) linkage of dihybrids
C) long overall distances (with respect to map distances)
D) linker of DNA
E) logarithm of odds
A) linkage over DNA
B) linkage of dihybrids
C) long overall distances (with respect to map distances)
D) linker of DNA
E) logarithm of odds

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
39
GGHH and ccdd individuals are crossed to each other and the F1 backcrossed to the gghh parent. The 4000 progeny included 1806 GgHh, 1794 gghh, 196 Gghh, and 204 ggHh individuals. GGhh and ggHH individuals are next crossed to each other and the F1 test-crossed to a gghh individual. A total of 1600 offspring appeared in the progeny. How many do you expect to be of GgHh genotype?
A) 40
B) 160
C) 320
D) 80
E) 1600
A) 40
B) 160
C) 320
D) 80
E) 1600

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
40
Why are the progeny of a testcross generally used to map loci? Why not the F2 progeny from an F1 × F1 cross?
A) Only recombinant offspring would be found in the progeny of an F1 × F1 cross.
B) The progeny of an F1 × F1 cross would be found in a 9:3:3:1 ratio when two genes are involved, whereas the progeny of a testcross would result in a 1:1:1:1 ratio.
C) It is easier to classify recombinant and parental offspring of a testcross than with the progeny of an F1 × F1 cross.
D) In a testcross more of the progeny would be expected to display the dominant phenotype than in the progeny of an F1 × F1 cross.
E) A testcross is more useful for mapping genes that are located near each other but, when genes are quite far apart on the same chromosome, an F1 × F1 cross actually is more useful.
A) Only recombinant offspring would be found in the progeny of an F1 × F1 cross.
B) The progeny of an F1 × F1 cross would be found in a 9:3:3:1 ratio when two genes are involved, whereas the progeny of a testcross would result in a 1:1:1:1 ratio.
C) It is easier to classify recombinant and parental offspring of a testcross than with the progeny of an F1 × F1 cross.
D) In a testcross more of the progeny would be expected to display the dominant phenotype than in the progeny of an F1 × F1 cross.
E) A testcross is more useful for mapping genes that are located near each other but, when genes are quite far apart on the same chromosome, an F1 × F1 cross actually is more useful.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
41
A physical map often measures _____, whereas a genetic map measures _____.
A) distances between chromosomes; distances between genes
B) map units between genes; physical distances along the chromosome
C) centiMorgans; base pairs
D) distances in base pairs along the chromosome; centiMorgans
E) map units between genes; centiMorgans
A) distances between chromosomes; distances between genes
B) map units between genes; physical distances along the chromosome
C) centiMorgans; base pairs
D) distances in base pairs along the chromosome; centiMorgans
E) map units between genes; centiMorgans

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
42
Three-factor testcrosses are only informative in gene mapping when which of the following occurs?
A) One parent is homozygous recessive for the three genes, and the other parent is homozygous dominant.
B) All three genes are located on separate chromosomes, and one parent is homozygous dominant for at least two of these genes.
C) Both parents are homozygous for the three genes.
D) One parent is heterozygous for the three genes, and the other parent is homozygous recessive.
E) One of the genes must be located on a sex chromosome and be heterozygous, and the other two genes must be located on an autosome and be homozygous.
A) One parent is homozygous recessive for the three genes, and the other parent is homozygous dominant.
B) All three genes are located on separate chromosomes, and one parent is homozygous dominant for at least two of these genes.
C) Both parents are homozygous for the three genes.
D) One parent is heterozygous for the three genes, and the other parent is homozygous recessive.
E) One of the genes must be located on a sex chromosome and be heterozygous, and the other two genes must be located on an autosome and be homozygous.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
43
Linked genes are:
A) allelic.
B) dominant.
C) on different chromosomes.
D) on the same chromosome.
E) recessive lethal.
A) allelic.
B) dominant.
C) on different chromosomes.
D) on the same chromosome.
E) recessive lethal.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
44
A study is done on families in Sweden that are segregating a genetic disorder. Lod-score analysis indicates that the gene involved in the disorder shows a strong likelihood of linkage with a particular DNA marker locus. However, a second study done in Italy with other families segregating the same genetic disorder results in lod-score values that strongly indicate the lack of linkage between the gene and the same DNA marker locus. Assuming that both studies were performed appropriately, what is the most likely explanation for the different outcomes?
A) No recombinants were found in the families studied in Sweden.
B) The allele that caused the disorder was in coupling linkage with one of the DNA marker alleles in the Swedish families but was in repulsion linkage in the Italian families.
C) This disorder is caused by mutations in either of two different genes; one of these genes is linked to the DNA marker locus and the other gene is not.
D) In the Italian families, the gene involved with the disorder is near a lethal allele at another locus and most of the parental or nonrecombinant genotypes contain the lethal allele; this reduces the number of nonrecombinants observed.
E) Linkage should have been observed in the Italian families, but there were only two alleles at the DNA marker locus that prevented recombinant offspring from appearing.
A) No recombinants were found in the families studied in Sweden.
B) The allele that caused the disorder was in coupling linkage with one of the DNA marker alleles in the Swedish families but was in repulsion linkage in the Italian families.
C) This disorder is caused by mutations in either of two different genes; one of these genes is linked to the DNA marker locus and the other gene is not.
D) In the Italian families, the gene involved with the disorder is near a lethal allele at another locus and most of the parental or nonrecombinant genotypes contain the lethal allele; this reduces the number of nonrecombinants observed.
E) Linkage should have been observed in the Italian families, but there were only two alleles at the DNA marker locus that prevented recombinant offspring from appearing.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
45
The results of linkage analysis for DNA marker A and the p53 gene are shown below. What is the best estimate for the approximate genetic distance between marker A and the p53 gene in humans?
A) 1 m.u.
B) 5 m.u.
C) 10 m.u.
D) 20 m.u.
E) 30 m.u.
A) 1 m.u.
B) 5 m.u.
C) 10 m.u.
D) 20 m.u.
E) 30 m.u.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
46
In addition to determining genotypes, two- and three-factor testcrosses can be used to:
A) map gene loci.
B) screen recessive mutants.
C) measure heritability.
D) determine parental origin.
E) determine the physical location of genes.
A) map gene loci.
B) screen recessive mutants.
C) measure heritability.
D) determine parental origin.
E) determine the physical location of genes.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
47
The map distances for genes that are close to each other are more accurate than map distances for genes that are quite far apart because:
A) with genes that are far apart, double crossovers and other multiple-crossover events often lead to lethal recombinants that reduce the number of recombinant progeny.
B) with genes that are far apart, double crossovers and other multiple-crossover events often lead to nonrecombinant or parental offspring and thus reduce the true map distance.
C) crossover interference will cause more double crossovers and other multiple-crossover events to occur than would be expected and thus result in a higher number of recombinant progeny than expected to occur with genes that are far apart.
D) double crossovers and other multiple-crossover events occur more often when genes are close to each other and can be readily detected, so these map distances are more accurate than those for genes that are far apart.
E) when genes are far apart, single-crossover recombinant classes are more difficult to detect than when genes are close together.
A) with genes that are far apart, double crossovers and other multiple-crossover events often lead to lethal recombinants that reduce the number of recombinant progeny.
B) with genes that are far apart, double crossovers and other multiple-crossover events often lead to nonrecombinant or parental offspring and thus reduce the true map distance.
C) crossover interference will cause more double crossovers and other multiple-crossover events to occur than would be expected and thus result in a higher number of recombinant progeny than expected to occur with genes that are far apart.
D) double crossovers and other multiple-crossover events occur more often when genes are close to each other and can be readily detected, so these map distances are more accurate than those for genes that are far apart.
E) when genes are far apart, single-crossover recombinant classes are more difficult to detect than when genes are close together.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
48
Consider the following three-factor (trihybrid) testcross: 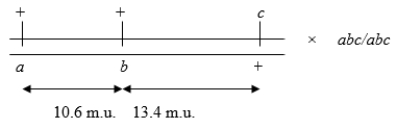 Calculate the number of individuals of a+a bb c+c genotype if 1000 progeny result from this testcross.
Calculate the number of individuals of a+a bb c+c genotype if 1000 progeny result from this testcross.
A) about 102
B) about 46
C) about 130
D) about 65
E) about 250
 Calculate the number of individuals of a+a bb c+c genotype if 1000 progeny result from this testcross.
Calculate the number of individuals of a+a bb c+c genotype if 1000 progeny result from this testcross.A) about 102
B) about 46
C) about 130
D) about 65
E) about 250

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
49
A low coefficient of coincidence indicates that:
A) far fewer double-crossover recombinant progeny were recovered from a testcross than would be expected from the map distances of the genes involved.
B) crossing over has been enhanced for genes that are located near the centromere of chromosomes because there is less interference of one crossover on the occurrence of a second crossover event.
C) single-crossover recombinant classes in the progeny have been increased because the genes involved produce lethal phenotypes when in parental gene combinations.
D) there is a large map distance between one of the outside genes in the heterozygous parent and the middle gene, while there is a short map distance between the middle gene and the other outside gene.
E) the physical distance between two genes is very short compared with the genetic map distance between these two genes.
A) far fewer double-crossover recombinant progeny were recovered from a testcross than would be expected from the map distances of the genes involved.
B) crossing over has been enhanced for genes that are located near the centromere of chromosomes because there is less interference of one crossover on the occurrence of a second crossover event.
C) single-crossover recombinant classes in the progeny have been increased because the genes involved produce lethal phenotypes when in parental gene combinations.
D) there is a large map distance between one of the outside genes in the heterozygous parent and the middle gene, while there is a short map distance between the middle gene and the other outside gene.
E) the physical distance between two genes is very short compared with the genetic map distance between these two genes.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
50
If a three-strand double crossover occurs between two genes during meiosis, what is the expected outcome with respect to these two genes?
A) Two chromatids are parental and two chromatids are recombinant.
B) All four chromatid are parental.
C) All four chromatids are recombinant.
D) Three chromatids are recombinant and one is parental.
E) Three chromatids are parental and one is recombinant.
A) Two chromatids are parental and two chromatids are recombinant.
B) All four chromatid are parental.
C) All four chromatids are recombinant.
D) Three chromatids are recombinant and one is parental.
E) Three chromatids are parental and one is recombinant.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
51
In maize (corn), assume that the genes A and B are linked and 30 map units apart. If a plant of Ab/aB is selfed, what proportion of the progeny would be expected to be of ab/ab genotype?
A) 2.25%
B) 15%
C) 9%
D) 30%
E) 4.5%
A) 2.25%
B) 15%
C) 9%
D) 30%
E) 4.5%

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
52
Interference occurs when:
A) two genes are assorting independently.
B) two genes are far apart on a genetic map.
C) one crossover inhibits another.
D) the number of recombinant progeny classes in the testcross of a heterozygote exceeds the number of parental progeny.
E) a crossover causes the termination of the meiosis event in which the crossover is occurring.
A) two genes are assorting independently.
B) two genes are far apart on a genetic map.
C) one crossover inhibits another.
D) the number of recombinant progeny classes in the testcross of a heterozygote exceeds the number of parental progeny.
E) a crossover causes the termination of the meiosis event in which the crossover is occurring.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
53
A situation in which the coefficient of coincidence is greater than 1.0 would indicate that:
A) the interference is high and one crossover suppresses the occurrence of a second one.
B) no double crossovers were found in the progeny of a testcross, even though some were expected based on probability.
C) double crossovers were found in the progeny of a testcross, but there were fewer of them than would be expected based on probability.
D) there were more double crossovers in the progeny than would be expected based on probability.
E) the genes involved were actually assorting independently.
A) the interference is high and one crossover suppresses the occurrence of a second one.
B) no double crossovers were found in the progeny of a testcross, even though some were expected based on probability.
C) double crossovers were found in the progeny of a testcross, but there were fewer of them than would be expected based on probability.
D) there were more double crossovers in the progeny than would be expected based on probability.
E) the genes involved were actually assorting independently.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
54
What is a major difference in using lod-score analysis compared to using association studies in determining gene locations in humans?
A) Lod-score analysis relies on family or pedigree data, while association studies use population data.
B) Lod-score analysis requires that the loci being mapped must be on different chromosome arms, while association studies can map genes on different chromosomes.
C) Association studies compare genotypes between parents and their children, while lod-score analysis compares genotypes between siblings of the same family.
D) Lod-score analysis requires isolated human populations, while association studies require very large family pedigrees.
E) Lod-score analysis requires a large number of genes with multiple alleles, while association studies can use genes that have only two alleles.
A) Lod-score analysis relies on family or pedigree data, while association studies use population data.
B) Lod-score analysis requires that the loci being mapped must be on different chromosome arms, while association studies can map genes on different chromosomes.
C) Association studies compare genotypes between parents and their children, while lod-score analysis compares genotypes between siblings of the same family.
D) Lod-score analysis requires isolated human populations, while association studies require very large family pedigrees.
E) Lod-score analysis requires a large number of genes with multiple alleles, while association studies can use genes that have only two alleles.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
55
In a two-point linkage analysis, genes a and b have been found to be 26 m.u. apart on the same chromosome. A third gene, c, has just been discovered and found to be located between a and b. A three-point linkage analysis with a, b, and c indicates that a and b are actually 33 m.u. apart rather than 26 m.u. Why does the three-point analysis give a different map distance for a and b than does the two-point linkage analysis, and which is more accurate?

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
56
Consider the following three-point (trihybrid) testcross: 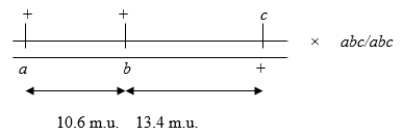 Calculate the number of double crossovers you would expect to observe if 1000 progeny result from this testcross assuming a coefficient of coincidence of 0.25.
Calculate the number of double crossovers you would expect to observe if 1000 progeny result from this testcross assuming a coefficient of coincidence of 0.25.
A) about 14
B) about 26
C) about 10
D) about 4
E) none because of crossover interference
 Calculate the number of double crossovers you would expect to observe if 1000 progeny result from this testcross assuming a coefficient of coincidence of 0.25.
Calculate the number of double crossovers you would expect to observe if 1000 progeny result from this testcross assuming a coefficient of coincidence of 0.25.A) about 14
B) about 26
C) about 10
D) about 4
E) none because of crossover interference

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
57
You are studying three genes X, Y, and Z that are linked (in that order) in the Imperial Scorpion Pandinus imperator. The distance between X and Y is 10 m.u., the distance between Y and Z is 8 m.u. You conduct a testcross by crossing a heterozygous female with a homozygous recessive male and obtain 1500 testcross progeny. When the progeny are analyzed, you find five double-crossover offspring. What is the interference value shown by this cross?
A) 0.008
B)0) 42
C) 0.12
D) 0.58
E) 0.22
A) 0.008
B)0) 42
C) 0.12
D) 0.58
E) 0.22

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
58
A testcross is performed on an individual to examine three linked genes. The most frequent phenotypes of the progeny were Abc and aBC, and the least frequent phenotypes were abc and ABC. What was the genotype of the heterozygous individual that is test-crossed with the correct order of the three genes?
A) Abc aBC
B) BAC/bac
C) bcA/BCa
D) aBc/AbC
E) bAc/BaC
A) Abc aBC
B) BAC/bac
C) bcA/BCa
D) aBc/AbC
E) bAc/BaC

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
59
Linkage disequilibrium is defined by which of the following?
A) positions in the genome where people vary by a single nucleotide base
B) the probability that two genes are linked
C) the nonrandom association between alleles in a haplotype
D) the occurrence of two alleles in the repulsion configuration
E) crossing over that occurs between two genes that are located close to each other
A) positions in the genome where people vary by a single nucleotide base
B) the probability that two genes are linked
C) the nonrandom association between alleles in a haplotype
D) the occurrence of two alleles in the repulsion configuration
E) crossing over that occurs between two genes that are located close to each other

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
60
A cell possessing two nuclei derived from different cells through cell fusion is called:
A) a heterokaryon.
B) a haplotype.
C) recombinant.
D) nonrecombinant.
E) None of the answers is correct.
A) a heterokaryon.
B) a haplotype.
C) recombinant.
D) nonrecombinant.
E) None of the answers is correct.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
61
You and a colleague are working on a rare Peruvian llama that appears to be susceptible to diabetes, a disease related to insulin function. Your colleague has established a somatic-cell hybrid panel, and you would like to figure out to which llama chromosome the gene that encodes the llama insulin receptor maps. You also have an assay that allows you to detect which of the somatic-cell lines can produce the insulin receptor. You assay the colleague's somatic-cell hybrid panel and get the following results. Which of the 74 llama chromosomes is the gene on?
A) 55
B) 7
C) 41
D) 68
E) 18
A) 55
B) 7
C) 41
D) 68
E) 18

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
62
Compared with a physical map, a genetic map:
A) is more accurate.
B) is less accurate.
C) is equally accurate.
D) measures different things.
E) cannot be made for humans.
A) is more accurate.
B) is less accurate.
C) is equally accurate.
D) measures different things.
E) cannot be made for humans.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
63
In using somatic-cell hybridization experiments, a human gene was found to be located on chromosome 6. However, when lod-score analysis was done to detect linkage between this gene and a DNA marker locus also known to be on chromosome 6, no linkage could be found between the marker locus and the gene. What is the MOST likely explanation for this result?
A) Somatic-cell hybridization experiments are not very accurate, and the gene may be on chromosome 5 or chromosome 7 instead of chromosome 6.
B) Too few recombinants could be found to indicate linkage in the lod-score analysis.
C) A lod-score analysis cannot be used when a DNA marker locus needs to mapped with respect to a gene locus.
D) The gene and the DNA marker locus are so far apart on chromosome 6 that they assort independently.
E) There were probably too few double-crossover events occurring between the two loci, so the lod score could not be determined accurately.
A) Somatic-cell hybridization experiments are not very accurate, and the gene may be on chromosome 5 or chromosome 7 instead of chromosome 6.
B) Too few recombinants could be found to indicate linkage in the lod-score analysis.
C) A lod-score analysis cannot be used when a DNA marker locus needs to mapped with respect to a gene locus.
D) The gene and the DNA marker locus are so far apart on chromosome 6 that they assort independently.
E) There were probably too few double-crossover events occurring between the two loci, so the lod score could not be determined accurately.

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
64
Assume that you discover a new human gene that you believe is located on the Y chromosome although not in the region (pseudoautosomal) of the Y that is homologous with part of the X chromosome. How would you map this gene with respect to the other genes on the Y chromosome?

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck
65
A geneticist finds that a human gene and a particular DNA marker locus are located on chromosome 8 on the basis of somatic-cell hybridization studies. However, when lod-score analysis is done with these two loci using family pedigrees, no evidence for linkage between the two loci can be found. Assuming that both types of studies were done correctly and the results are valid, how would you explain the different outcomes?

Unlock Deck
Unlock for access to all 65 flashcards in this deck.
Unlock Deck
k this deck


