Deck 19: Molecular Genetic Analysis and Biotechnology
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/72
Play
Full screen (f)
Deck 19: Molecular Genetic Analysis and Biotechnology
1
Antibodies are to Western blots as _____ is/are to Southern blots.
A) RNA probes
B) proteins
C) DNA probes
D) amino acids
E) DNA or RNA probes
A) RNA probes
B) proteins
C) DNA probes
D) amino acids
E) DNA or RNA probes
E
2
A gene does not contain the necessary restriction enzyme sites for cloning into a plasmid vector. What is a possible option?
A) Increase the amount of gene used.
B) Add linkers to generate new restriction enzyme sites.
C) Use a cosmid as a cloning vector.
D) Use dideoxy sequencing to obtain the sequence of the gene.
E) Cut the DNA with a blunt end cutter.
A) Increase the amount of gene used.
B) Add linkers to generate new restriction enzyme sites.
C) Use a cosmid as a cloning vector.
D) Use dideoxy sequencing to obtain the sequence of the gene.
E) Cut the DNA with a blunt end cutter.
B
3
A scientist edits a mammalian genome by inserting a GFP reporter sequence downstream of a gene of interest. Which of the following methods is she likely to have used to make this change?
A) site-directed mutagenesis
B) oligonucleotide-directed mutagenesis
C) CRISPR-Cas9 genome editing followed by nonhomologous end joining (NHEJ)
D) CRISPR-Cas9 genome editing followed by homologous recombination (HR) with a donor template
E) mutagenesis with radiation
A) site-directed mutagenesis
B) oligonucleotide-directed mutagenesis
C) CRISPR-Cas9 genome editing followed by nonhomologous end joining (NHEJ)
D) CRISPR-Cas9 genome editing followed by homologous recombination (HR) with a donor template
E) mutagenesis with radiation
D
4
Southern blotting is a technique used to transfer _____ to a solid Moderate.
A) DNA
B) RNA
C) protein
D) DNA and RNA
E) DNA, RNA, or protein
A) DNA
B) RNA
C) protein
D) DNA and RNA
E) DNA, RNA, or protein

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
5
During gel electrophoresis, large DNA fragments will _____ small DNA fragments.
A) migrate more rapidly than
B) migrate at the same speed as
C) migrate more slowly than
D) cause degradation of
E) separate into
A) migrate more rapidly than
B) migrate at the same speed as
C) migrate more slowly than
D) cause degradation of
E) separate into

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
6
All of the following are requirements of a bacterial cloning vector EXCEPT:
A) origin of replication.
B) unique restriction enzyme sites.
C) Ti plasmid.
D) selectable markers.
A) origin of replication.
B) unique restriction enzyme sites.
C) Ti plasmid.
D) selectable markers.

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
7
Which of the following statements CORRECTLY describes DNA ligase?
A) DNA ligase forms hydrogen bonds between nucleotide bases.
B) DNA ligase can seal nicks between amino acids.
C) DNA ligase recognizes and cuts at specific sequences.
D) DNA ligase is necessary for creating recombinant plasmids.
E) DNA ligase is a requirement of a sequencing reaction.
A) DNA ligase forms hydrogen bonds between nucleotide bases.
B) DNA ligase can seal nicks between amino acids.
C) DNA ligase recognizes and cuts at specific sequences.
D) DNA ligase is necessary for creating recombinant plasmids.
E) DNA ligase is a requirement of a sequencing reaction.

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
8
You are interested in a particular segment of rhinoceros DNA and would like to clone it into a cloning plasmid. You have the following restriction map of the region that includes the DNA of interest and the plasmid (E = EcoRI, H = HindIII, X = XbaI, S = SphI, N = NotI). 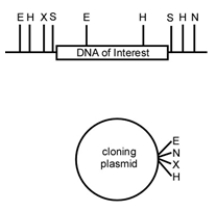 Which restriction enzymes would you choose to clone the DNA of interest into the cloning vector?
Which restriction enzymes would you choose to clone the DNA of interest into the cloning vector?
A) E and H
B) S
C) X and N
D) S and N
 Which restriction enzymes would you choose to clone the DNA of interest into the cloning vector?
Which restriction enzymes would you choose to clone the DNA of interest into the cloning vector?A) E and H
B) S
C) X and N
D) S and N

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
9
Which of the following traits of type I restriction enzymes make them unsuitable for recombinant DNA technology? (Select all that apply.)
A) They are large, multi-subunit enzymes.
B) They make double-stranded cuts in DNA.
C) They cleave and methylate DNA.
D) They cleave at sequences far from their recognition site.
E) They were discovered in bacteria.
A) They are large, multi-subunit enzymes.
B) They make double-stranded cuts in DNA.
C) They cleave and methylate DNA.
D) They cleave at sequences far from their recognition site.
E) They were discovered in bacteria.

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
10
Which of the following is a set of molecular techniques for locating, isolating, altering, and studying DNA segments?
A) in situ hybridization
B) gel electrophoresis
C) molecular cloning
D) Southern blotting
E) recombinant DNA technology
A) in situ hybridization
B) gel electrophoresis
C) molecular cloning
D) Southern blotting
E) recombinant DNA technology

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
11
Which of the following statements does NOT describe a challenge of working at the molecular level?
A) Cells contain thousands of genes.
B) Individual genes cannot be seen.
C) It is not possible to isolate DNA in a stable form.
D) A genome can consist of billions of base pairs.
E) No physical features mark the beginning or end of a gene.
A) Cells contain thousands of genes.
B) Individual genes cannot be seen.
C) It is not possible to isolate DNA in a stable form.
D) A genome can consist of billions of base pairs.
E) No physical features mark the beginning or end of a gene.

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
12
Gel electrophoresis can be used to separate DNA on the basis of:
A) size.
B) electrical charge.
C) nucleotide content.
D) the probe used.
E) both size and electrical charge.
A) size.
B) electrical charge.
C) nucleotide content.
D) the probe used.
E) both size and electrical charge.

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
13
The recognition site for BamHI is 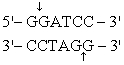
) Which of the following restriction sites when digested would create a cohesive end that could be ligated to a BamHI digested DNA fragment?
A) EcoRII
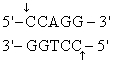
B) PvuII
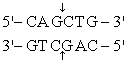
C) EcoRI
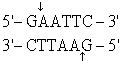
D) BglII
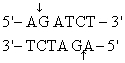
E) CofI
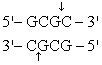
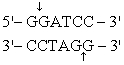
) Which of the following restriction sites when digested would create a cohesive end that could be ligated to a BamHI digested DNA fragment?
A) EcoRII
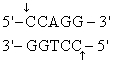
B) PvuII
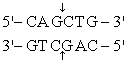
C) EcoRI
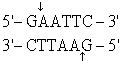
D) BglII
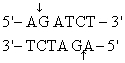
E) CofI
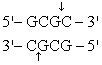

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
14
What are Northern analyses used for? Describe the steps involved in performing a Northern analysis, and describe how levels of gene expression are determined.

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
15
In the introduction to this chapter, the use of the CRISPR-Cas9 system to make a deletion of exon 23 containing a premature stop codon in mdx mice was described. The resulting mice had partially restored dystrophin protein and muscle function. Which of the following CRISRP-Cas9 mediated changes might lead to a better restoration of dystrophin protein and muscle function?
A) A deletion of exon 24 in addition to exon 23
B) A deletion of intron 23
C) A deletion of 30 bases within exon 23
D) A deletion of 25 bases within exon 21
E) An insertion of a corrected copy of exon 23 after the mutant exon 23
A) A deletion of exon 24 in addition to exon 23
B) A deletion of intron 23
C) A deletion of 30 bases within exon 23
D) A deletion of 25 bases within exon 21
E) An insertion of a corrected copy of exon 23 after the mutant exon 23

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
16
The haploid human genome contains about 3 × 109 nucleotides. On average, how many DNA fragments would be produced if this DNA was digested with restriction enzyme PstI (a 6-base cutter)? RsaI (a 4-base cutter)? How often would an 8-base cutter cleave?

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
17
A scientist is studying a normal tissue sample and a cancerous tissue sample. What method might she use to determine whether the transcription of gene X is upregulated in the cancerous tissue sample?
A) Carrying out a Southern blot using the cancerous tissue sample only
B) Carrying out a Southern blot using both samples
C) Carrying out a Northern blot using the cancerous tissue sample only
D) Carrying out a Northern blot using both samples
E) Carrying out a Western blot using the cancerous tissue sample only
A) Carrying out a Southern blot using the cancerous tissue sample only
B) Carrying out a Southern blot using both samples
C) Carrying out a Northern blot using the cancerous tissue sample only
D) Carrying out a Northern blot using both samples
E) Carrying out a Western blot using the cancerous tissue sample only

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
18
Which of the following would be MOST appropriate for cloning a gene that is 300 kb in size?
A) plasmid
B) cosmid
C) phage lambda
D) BAC
E) yeast phage
A) plasmid
B) cosmid
C) phage lambda
D) BAC
E) yeast phage

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
19
A scientist attempts to use the CRISPR-Cas9 system to edit a single gene within a cell. She finds that, in addition to editing the desired gene, she has edited several other genes as well. What are reasonable options for her to try next in order to target just the gene of interest? (Select all that apply.)
A) Add less Cas9 enzyme.
B) Create a longer sgRNA.
C) Try a different sgRNA that will still pair within the gene of interest.
D) Use separate crRNA and tracrRNA molecules.
E) Use a higher-fidelity version of Cas9.
A) Add less Cas9 enzyme.
B) Create a longer sgRNA.
C) Try a different sgRNA that will still pair within the gene of interest.
D) Use separate crRNA and tracrRNA molecules.
E) Use a higher-fidelity version of Cas9.

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
20
Which of the following statements is NOT correct regarding type II restriction enzymes?
A) They can create blunt ends.
B) They make double-stranded cuts in DNA.
C) They recognize specific sequences and make cuts further away from the recognition sequence.
D) They are named based on their bacterial origin.
A) They can create blunt ends.
B) They make double-stranded cuts in DNA.
C) They recognize specific sequences and make cuts further away from the recognition sequence.
D) They are named based on their bacterial origin.

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
21
Use the following to answer questions
Figure A below shows a restriction map of a rare prokaryotic gene with its direction of transcription indicated by the arrow. Figure B shows the unique restriction sites contained within a plasmid-cloning vector. The blackened region in Figure A represents the amino acid coding sequence of a protein that can be used in humans as a vaccine. The striped region in Figure B is a highly active, constitutive (unregulated) prokaryotic promoter region. Letters indicate the cleavage sites for different restriction enzymes. Known DNA sequences are indicated by short thick lines.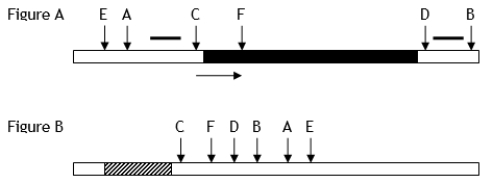
After trying to isolate and then insert the coding region (Figure
A) under the control of the indicated promoter in the cloning vector (Figure
B), you found that all the transformed bacterial cells contained either one of two smaller portions of the coding region for the gene, and some of the fragments were inserted backward (with regard to reading frame) into the cloning vector. How would you explain these observations?
Figure A below shows a restriction map of a rare prokaryotic gene with its direction of transcription indicated by the arrow. Figure B shows the unique restriction sites contained within a plasmid-cloning vector. The blackened region in Figure A represents the amino acid coding sequence of a protein that can be used in humans as a vaccine. The striped region in Figure B is a highly active, constitutive (unregulated) prokaryotic promoter region. Letters indicate the cleavage sites for different restriction enzymes. Known DNA sequences are indicated by short thick lines.

After trying to isolate and then insert the coding region (Figure
A) under the control of the indicated promoter in the cloning vector (Figure
B), you found that all the transformed bacterial cells contained either one of two smaller portions of the coding region for the gene, and some of the fragments were inserted backward (with regard to reading frame) into the cloning vector. How would you explain these observations?

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
22
You are examining a 1000-bp DNA fragment as part of a forensic analysis. If the limit of detection of this molecule is 9 × 108 molecules, what is the minimum number of PCR cycles you would have to run to detect a PCR product generated from a single molecule of the template?

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
23
What is the purpose of Taq polymerase in a PCR reaction?
A) DNA denaturation
B) primer annealing
C) DNA synthesis
D) heating of the reaction
E) heating of the reaction and DNA denaturation
A) DNA denaturation
B) primer annealing
C) DNA synthesis
D) heating of the reaction
E) heating of the reaction and DNA denaturation

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
24
You have discovered a gene that enables organisms to accumulate gold in their tissues by concentrating trace amounts found in normal soil. You want to transfer this gene into a plant. Order the steps below that would accomplish this goal. 1. Infect the plant with the Agrobacterium strain.
2) Digest the gold gene and a Ti plasmid with appropriate restriction enzymes.
3) Insert the gold gene into the Ti plasmid.
4) Amplify the gold gene with PCR.
5) Transfer the recombinant Ti plasmid into Agrobacterium tumefaciens.
6) Use a selectable marker to identify plant cells that have integrated the recombinant plasmid into their genome.
A) 4, 2, 3, 5, 1, 6
B) 4, 5, 2, 3, 1, 6
C) 4, 2, 5, 1, 6, 3
D) 4, 3, 2, 5, 1, 6
2) Digest the gold gene and a Ti plasmid with appropriate restriction enzymes.
3) Insert the gold gene into the Ti plasmid.
4) Amplify the gold gene with PCR.
5) Transfer the recombinant Ti plasmid into Agrobacterium tumefaciens.
6) Use a selectable marker to identify plant cells that have integrated the recombinant plasmid into their genome.
A) 4, 2, 3, 5, 1, 6
B) 4, 5, 2, 3, 1, 6
C) 4, 2, 5, 1, 6, 3
D) 4, 3, 2, 5, 1, 6

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
25
Which of the following is NOT a bacterial cloning vector?
A) plasmid
B) bacteriophage
C) agrobacterium
D) bacterial artificial chromosome
E) cosmid
A) plasmid
B) bacteriophage
C) agrobacterium
D) bacterial artificial chromosome
E) cosmid

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
26
Consider a tobacco plant cell that was able to express a toxin that was lethal to insects and was resistant to the antibiotic kanamycin. Which of the following genes would this cell contain?
A) Bt
B) neo+
C) lacZ
D) Bt and neo+ only
E) Bt, neo+, and lacZ
A) Bt
B) neo+
C) lacZ
D) Bt and neo+ only
E) Bt, neo+, and lacZ

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
27
Use the following to answer questions
A scientist carries out a ligation reaction designed to insert a foreign piece of DNA into a plasmid that contains the front end of the lacZ gene within the multiple cloning site (MCS). He carries out three transformations in parallel with lacZ- bacteria. The three transformations (i, ii, and iii) contain the following:
i. sterile water
ii. a sample of the original plasmid
iii. the ligation reaction
He then plates the transformations on medium containing X-gal and ampicillin.
What results would indicate the scientist should proceed to culture colonies from (iii) that might contain the desired clone?
A) Many blue colonies on all three plates.
B) White colonies on plates (i) and (ii) and predominantly blue colonies on plate (iii).
C) No colonies on plate (i), white colonies on plate (ii), and predominantly blue colonies on plate (iii).
D) No colonies on plate (i), blue colonies on plate (ii), and predominantly white colonies on plate (iii).
E) No colonies on plate (i), blue colonies on plate (ii), and predominantly blue colonies on plate (iii).
A scientist carries out a ligation reaction designed to insert a foreign piece of DNA into a plasmid that contains the front end of the lacZ gene within the multiple cloning site (MCS). He carries out three transformations in parallel with lacZ- bacteria. The three transformations (i, ii, and iii) contain the following:
i. sterile water
ii. a sample of the original plasmid
iii. the ligation reaction
He then plates the transformations on medium containing X-gal and ampicillin.
What results would indicate the scientist should proceed to culture colonies from (iii) that might contain the desired clone?
A) Many blue colonies on all three plates.
B) White colonies on plates (i) and (ii) and predominantly blue colonies on plate (iii).
C) No colonies on plate (i), white colonies on plate (ii), and predominantly blue colonies on plate (iii).
D) No colonies on plate (i), blue colonies on plate (ii), and predominantly white colonies on plate (iii).
E) No colonies on plate (i), blue colonies on plate (ii), and predominantly blue colonies on plate (iii).

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
28
Use the following to answer questions
A student carries out PCR using the following steps:
Step 1: 94°C for 1 minute
Step 2: 60°C for 30 seconds
Step 3: 72°C for 30 seconds
After subjecting his PCR reaction to gel electrophoresis, the student sees no PCR product on the gel. What error(s) might he have made? (Select all that apply.)
A) He carried out step 1 at too low a temperature.
B) He carried out step 1 at too high a temperature.
C) He carried out step 2 at too low a temperature.
D) He carried out step 2 at too high a temperature.
E) He carried out step 3 for too short a time.
A student carries out PCR using the following steps:
Step 1: 94°C for 1 minute
Step 2: 60°C for 30 seconds
Step 3: 72°C for 30 seconds
After subjecting his PCR reaction to gel electrophoresis, the student sees no PCR product on the gel. What error(s) might he have made? (Select all that apply.)
A) He carried out step 1 at too low a temperature.
B) He carried out step 1 at too high a temperature.
C) He carried out step 2 at too low a temperature.
D) He carried out step 2 at too high a temperature.
E) He carried out step 3 for too short a time.

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
29
Which of the following is NOT a potential benefit of using transgenic plants?
A) They can reduce the use of harmful chemical pesticides in the United States and thus provide an ecological benefit.
B) They can generate restriction enzyme sites on a foreign gene of interest to be cloned.
C) They often increase yields, providing more food per acre and reducing the amount of land needed for agricultural use.
D) They can allow crops to be grown on land previously unavailable for productive agricultural use.
E) They can be used to express large quantities of specific biological products more cheaply and quickly than by expression in animal systems.
A) They can reduce the use of harmful chemical pesticides in the United States and thus provide an ecological benefit.
B) They can generate restriction enzyme sites on a foreign gene of interest to be cloned.
C) They often increase yields, providing more food per acre and reducing the amount of land needed for agricultural use.
D) They can allow crops to be grown on land previously unavailable for productive agricultural use.
E) They can be used to express large quantities of specific biological products more cheaply and quickly than by expression in animal systems.

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
30
Use the following to answer questions
A scientist carries out a ligation reaction designed to insert a foreign piece of DNA into a plasmid that contains the front end of the lacZ gene within the multiple cloning site (MCS). He carries out three transformations in parallel with lacZ- bacteria. The three transformations (i, ii, and iii) contain the following:
i. sterile water
ii. a sample of the original plasmid
iii. the ligation reaction
He then plates the transformations on medium containing X-gal and ampicillin.
The scientist observed colonies on plate (i). Which of the following might be reasons(s) for this? (Select all that apply.)
A) The plasmid he used has a kanamycin resistance marker instead of an ampicillin resistance marker.
B) He transformed cells that were not actually ampicillin sensitive.
C) Too high a concentration of ampicillin was added to the plates.
D) Too low a concentration of ampicillin was added to the plates.
E) The water sample was contaminated with the original plasmid.
A scientist carries out a ligation reaction designed to insert a foreign piece of DNA into a plasmid that contains the front end of the lacZ gene within the multiple cloning site (MCS). He carries out three transformations in parallel with lacZ- bacteria. The three transformations (i, ii, and iii) contain the following:
i. sterile water
ii. a sample of the original plasmid
iii. the ligation reaction
He then plates the transformations on medium containing X-gal and ampicillin.
The scientist observed colonies on plate (i). Which of the following might be reasons(s) for this? (Select all that apply.)
A) The plasmid he used has a kanamycin resistance marker instead of an ampicillin resistance marker.
B) He transformed cells that were not actually ampicillin sensitive.
C) Too high a concentration of ampicillin was added to the plates.
D) Too low a concentration of ampicillin was added to the plates.
E) The water sample was contaminated with the original plasmid.

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
31
Which of the following statements is NOT true regarding the basic components required for a bacterial cloning vector?
A) Selectable markers provide a means for preferentially allowing growth of only those bacterial cells that have been transformed with the cloning vector.
B) Unique restriction enzyme sites allow for larger pieces of foreign DNA to be inserted into the bacterial cloning vector.
C) Unique restriction enzyme sites provide a means for inserting the foreign DNA into the cloning vector at a specific known sequence site.
D) A bacterial origin of replication ensures that the plasmid is replicated while present within the bacterial cell.
E) Selectable markers provide a means for selecting cells that have been transformed with a recombinant plasmid.
A) Selectable markers provide a means for preferentially allowing growth of only those bacterial cells that have been transformed with the cloning vector.
B) Unique restriction enzyme sites allow for larger pieces of foreign DNA to be inserted into the bacterial cloning vector.
C) Unique restriction enzyme sites provide a means for inserting the foreign DNA into the cloning vector at a specific known sequence site.
D) A bacterial origin of replication ensures that the plasmid is replicated while present within the bacterial cell.
E) Selectable markers provide a means for selecting cells that have been transformed with a recombinant plasmid.

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
32
Which of the following represents an appropriate cloning vector for cloning a gene into a bacterial cell?
A) YAC
B) Ti plasmid
C) plasmid
D) Agrobacterium tumefaciens
E) lacZ
A) YAC
B) Ti plasmid
C) plasmid
D) Agrobacterium tumefaciens
E) lacZ

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
33
The difference between PCR and real-time PCR is that real-time PCR:
A) can measure the amount of DNA amplified as the reaction proceeds, while standard PCR cannot.
B) can amplify DNA a billion-fold within just a few hours, while standard PCR cannot.
C) can determine the DNA sequence, while standard PCR cannot.
D) uses DNA polymerase, while standard PCR does not.
E) requires primers, while standard PCR does not.
A) can measure the amount of DNA amplified as the reaction proceeds, while standard PCR cannot.
B) can amplify DNA a billion-fold within just a few hours, while standard PCR cannot.
C) can determine the DNA sequence, while standard PCR cannot.
D) uses DNA polymerase, while standard PCR does not.
E) requires primers, while standard PCR does not.

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
34
Which of the following can be used for genetic engineering in plants?
A) Ti plasmid
B) Agrobacterium tumefaciens
C) selectable markers
D) Ti plasmids and Agrobacterium tumefaciens only
E) Ti plasmids, Agrobacterium tumefaciens, and selectable markers
A) Ti plasmid
B) Agrobacterium tumefaciens
C) selectable markers
D) Ti plasmids and Agrobacterium tumefaciens only
E) Ti plasmids, Agrobacterium tumefaciens, and selectable markers

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
35
A cDNA encoding a protein that is specific for a particular strain (strain Q) of bacteria has been cloned. How would you determine whether an infection contains this particular bacterial strain?

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
36
Use the following to answer questions
A student carries out PCR using the following steps:
Step 1: 94°C for 1 minute
Step 2: 60°C for 30 seconds
Step 3: 72°C for 30 seconds
Which of the following lists the CORRECT terms for these three steps?
A) denaturation of the double-stranded template, extension of the new DNA molecules, primer annealing
B) denaturation of the double-stranded template, primer annealing, extension of the new DNA molecules
C) denaturation of the double-stranded template, extension of the new DNA molecules, hybridization of the template
D) degradation of the template, primer annealing, extension of the new DNA molecules
E) hybridization of the single-stranded templates, primer annealing, extension of the new DNA molecules
A student carries out PCR using the following steps:
Step 1: 94°C for 1 minute
Step 2: 60°C for 30 seconds
Step 3: 72°C for 30 seconds
Which of the following lists the CORRECT terms for these three steps?
A) denaturation of the double-stranded template, extension of the new DNA molecules, primer annealing
B) denaturation of the double-stranded template, primer annealing, extension of the new DNA molecules
C) denaturation of the double-stranded template, extension of the new DNA molecules, hybridization of the template
D) degradation of the template, primer annealing, extension of the new DNA molecules
E) hybridization of the single-stranded templates, primer annealing, extension of the new DNA molecules

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
37
Ampicillin resistance is to ampR as _____ is to lacZ.
A) Bt toxin
B) G418
C) cleavage of X-gal
D) penicillin resistance
E) gancyclovir
A) Bt toxin
B) G418
C) cleavage of X-gal
D) penicillin resistance
E) gancyclovir

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
38
Use the following to answer questions
Figure A below shows a restriction map of a rare prokaryotic gene with its direction of transcription indicated by the arrow. Figure B shows the unique restriction sites contained within a plasmid-cloning vector. The blackened region in Figure A represents the amino acid coding sequence of a protein that can be used in humans as a vaccine. The striped region in Figure B is a highly active, constitutive (unregulated) prokaryotic promoter region. Letters indicate the cleavage sites for different restriction enzymes. Known DNA sequences are indicated by short thick lines.
Explain how you would isolate and then insert the coding region (Figure
A) under the control of the indicated promoter in the cloning vector (Figure
B) to produce large amounts of the protein in bacterial cells. Assume that the cloning vector carries the gene for tetracycline (an antibiotic) resistance.
Figure A below shows a restriction map of a rare prokaryotic gene with its direction of transcription indicated by the arrow. Figure B shows the unique restriction sites contained within a plasmid-cloning vector. The blackened region in Figure A represents the amino acid coding sequence of a protein that can be used in humans as a vaccine. The striped region in Figure B is a highly active, constitutive (unregulated) prokaryotic promoter region. Letters indicate the cleavage sites for different restriction enzymes. Known DNA sequences are indicated by short thick lines.

Explain how you would isolate and then insert the coding region (Figure
A) under the control of the indicated promoter in the cloning vector (Figure
B) to produce large amounts of the protein in bacterial cells. Assume that the cloning vector carries the gene for tetracycline (an antibiotic) resistance.

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
39
Use the following to answer questions
A student carries out PCR using the following steps:
Step 1: 94°C for 1 minute
Step 2: 60°C for 30 seconds
Step 3: 72°C for 30 seconds
After electrophoresing his PCR reaction, the student sees the desired PCR product on the gel as well as several smaller bands. What error(s) might he have made? (Select all that apply.)
A) He carried out step 2 at too low a temperature
B) He carried out step 2 at too high a temperature.
C) He designed primers with repetitive sequences.
D) He contaminated the template DNA sample.
E) He carried out step 3 for too short a time.
A student carries out PCR using the following steps:
Step 1: 94°C for 1 minute
Step 2: 60°C for 30 seconds
Step 3: 72°C for 30 seconds
After electrophoresing his PCR reaction, the student sees the desired PCR product on the gel as well as several smaller bands. What error(s) might he have made? (Select all that apply.)
A) He carried out step 2 at too low a temperature
B) He carried out step 2 at too high a temperature.
C) He designed primers with repetitive sequences.
D) He contaminated the template DNA sample.
E) He carried out step 3 for too short a time.

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
40
You are attempting to determine whether a plant-derived processed food sample (for example, a corn chip) contains material from a genetically modified organism (GMO). First, you crush the sample and attempt to extract DNA from it. Next, you perform PCR using two different sets of primers. One primer set will amplify a DNA sequence present in all plants. The second primer set will amplify a DNA sequence only found in GMO plants.
a. Why must you use both sets of primers for this experiment?
b. In addition to the test sample, you obtain a negative control sample (food material you are certain does not contain GMO material) and a positive control sample (food material you are certain does contain GMO material). You perform the DNA extraction on these three samples and then the PCR reactions with each of the two primer sets described above. Complete the following table with your expectations for this PCR reaction.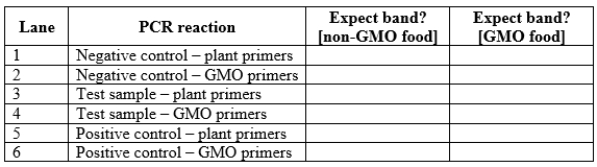 c. You obtain the results shown in the panel below. What conclusions can you draw from these reaction results? Does this test sample contain genetically modified components? Why or why not?
c. You obtain the results shown in the panel below. What conclusions can you draw from these reaction results? Does this test sample contain genetically modified components? Why or why not? 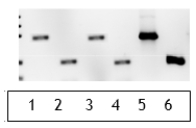
a. Why must you use both sets of primers for this experiment?
b. In addition to the test sample, you obtain a negative control sample (food material you are certain does not contain GMO material) and a positive control sample (food material you are certain does contain GMO material). You perform the DNA extraction on these three samples and then the PCR reactions with each of the two primer sets described above. Complete the following table with your expectations for this PCR reaction.
 c. You obtain the results shown in the panel below. What conclusions can you draw from these reaction results? Does this test sample contain genetically modified components? Why or why not?
c. You obtain the results shown in the panel below. What conclusions can you draw from these reaction results? Does this test sample contain genetically modified components? Why or why not? 

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
41
Suppose you have previously located and cloned a human gene on chromosome 2. You now believe there is an important disease-related gene nearby on the same chromosome. What technique might you employ to locate the disease-related gene?
A) chromosome walking
B) DNA fingerprinting
C) site-directed mutagenesis
D) RNAi
E) CRISPR-Cas genome editing
A) chromosome walking
B) DNA fingerprinting
C) site-directed mutagenesis
D) RNAi
E) CRISPR-Cas genome editing

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
42
Use the following to answer questions
You identify an RFLP in mice by digesting genomic DNA with HindIII enzyme and radiolabeling a piece of probe DNA. Southern blot analysis shows that the probe detects a 2-kb fragment in one strain of mice and a 4-kb fragment in another strain of mice. The two mice strains are crossed to produce an F1 generation. Two F1 siblings are mated to produce a dozen F2 progeny. You isolate genomic DNA from several F1 and F2 individuals, digest with HindIII, and perform a Southern blot with the same probe.
How many bands would be seen in the Southern blot of an F1 individual?
A) one
B) two
C) three
D) four
E) one or two
You identify an RFLP in mice by digesting genomic DNA with HindIII enzyme and radiolabeling a piece of probe DNA. Southern blot analysis shows that the probe detects a 2-kb fragment in one strain of mice and a 4-kb fragment in another strain of mice. The two mice strains are crossed to produce an F1 generation. Two F1 siblings are mated to produce a dozen F2 progeny. You isolate genomic DNA from several F1 and F2 individuals, digest with HindIII, and perform a Southern blot with the same probe.
How many bands would be seen in the Southern blot of an F1 individual?
A) one
B) two
C) three
D) four
E) one or two

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
43
Explain what an RFLP is. Why do RFLPs behave like "codominant" markers?

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
44
All of the following are required to create a cDNA library EXCEPT:
A) RNA polymerase.
B) DNA polymerase.
C) reverse transcriptase.
D) dNTPs.
E) mRNAs.
A) RNA polymerase.
B) DNA polymerase.
C) reverse transcriptase.
D) dNTPs.
E) mRNAs.

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
45
Which of the following is usually NOT a component used to generate cDNA?
A) reverse transcriptase
B) RNase
C) ligase
D) oligo(dT) chains
E) restriction enzymes
A) reverse transcriptase
B) RNase
C) ligase
D) oligo(dT) chains
E) restriction enzymes

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
46
You have two probes that detect RFLPs that are closely linked. You have been asked to help settle a paternity case. The two possible fathers have the following RFLP pedigree. The child and mother are also probed with these two RFLP probes. (The data below the pedigree are two Southern blots.) 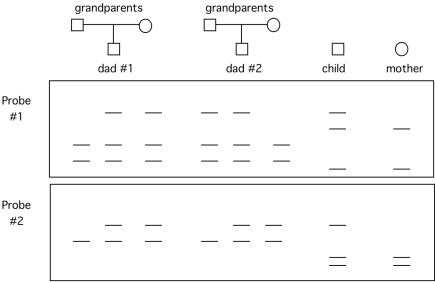 Which male is more likely to be the father? Explain your reasoning.
Which male is more likely to be the father? Explain your reasoning.
 Which male is more likely to be the father? Explain your reasoning.
Which male is more likely to be the father? Explain your reasoning.
Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
47
Which of the following DNA sequences are contained within a cDNA library?
A) exons
B) introns
C) promoters
D) enhancers
A) exons
B) introns
C) promoters
D) enhancers

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
48
You are handling a paternity lawsuit brought against five potential fathers by a woman. You isolated DNA from the mother, the child, and all the potential fathers. After using PCR to amplify specific polymorphic loci from each individual, you run a gel of the amplified products and stain with ethidium bromide to visualize the DNA fingerprints (shown below). Mo = mother; Ch = child; M1-M5 = potential fathers. B1-B10 indicate marker bands present in the child. Do these results suggest that any of the men could be the child's biological father? Explain your answer. 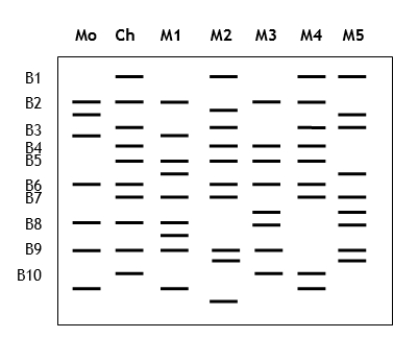


Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
49
One technique for finding a gene of interest involves first generating a genetic map to find the general location of the gene and then identifying the specific location of the gene. What is this technique called?
A) next-generation sequencing
B) DNA fingerprinting
C) positional cloning
D) in silico gene discovery
E) site-directed mutagenesis
A) next-generation sequencing
B) DNA fingerprinting
C) positional cloning
D) in silico gene discovery
E) site-directed mutagenesis

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
50
The lungfish Protopterus aethiopicus has a genome 38 times larger than that of humans. Most of the DNA in this species is noncoding repetitive DNA. What type of library would allow you to compare just the genes in lungfish to the genes in humans?
A) cDNA library
B) PCR library
C) genomic library
D) knockout library
E) transgenic library
A) cDNA library
B) PCR library
C) genomic library
D) knockout library
E) transgenic library

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
51
Which of the following statements is NOT correct?
A) Coding sequences for gene products can be isolated from cDNA libraries.
B) Antibodies are used for Northern blot analysis.
C) The number of STR copies is variable throughout human populations.
D) PCR amplification generates large numbers of linear DNA fragments.
E) RNA molecules can be used as hybridization probes in Southern blot analysis.
A) Coding sequences for gene products can be isolated from cDNA libraries.
B) Antibodies are used for Northern blot analysis.
C) The number of STR copies is variable throughout human populations.
D) PCR amplification generates large numbers of linear DNA fragments.
E) RNA molecules can be used as hybridization probes in Southern blot analysis.

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
52
Which of the following statements is INCORRECT regarding the differences between a genomic library and a cDNA library?
A) Genomic libraries contain fewer restriction enzyme sites, whereas cDNA libraries contain many more.
B) A genomic library is prepared from total genomic DNA, whereas a cDNA library is prepared from mRNA.
C) Genomic libraries contain much more sequence information and are much larger than cDNA libraries.
D) Genomic libraries contain coding and noncoding (regulatory, intron, etc.) sequences, whereas cDNA libraries contain only coding sequences along with their associated 5' and 3' untranslated regions.
E) cDNA libraries are generated with the use of reverse transcriptase, whereas genomic libraries are not.
A) Genomic libraries contain fewer restriction enzyme sites, whereas cDNA libraries contain many more.
B) A genomic library is prepared from total genomic DNA, whereas a cDNA library is prepared from mRNA.
C) Genomic libraries contain much more sequence information and are much larger than cDNA libraries.
D) Genomic libraries contain coding and noncoding (regulatory, intron, etc.) sequences, whereas cDNA libraries contain only coding sequences along with their associated 5' and 3' untranslated regions.
E) cDNA libraries are generated with the use of reverse transcriptase, whereas genomic libraries are not.

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
53
The full-length (i.e., containing the entire protein coding region) cDNA for a specific eukaryotic gene in humans is 1500 nucleotides long. You screen a pig genomic library with this cDNA and isolate two genomic clones of different lengths. Both clones are sequenced and found to be 1900 and 2100 nucleotides long from start codon to stop codon. How would you explain the presence of two genomic clones in pigs and the discrepancies in their lengths compared to the cDNA probe?

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
54
Use the following to answer questions
A fragment of DNA is cloned into a plasmid with a sequencing primer-binding site. After dideoxy sequencing, the gel pattern shown in this diagram is obtained.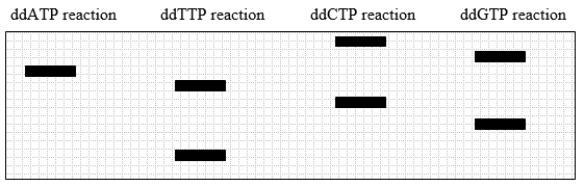
What was the sequence of the DNA strand that acted as the template in the sequencing reaction?
A) 5' GCTAGCA 3'
B) 5' ACGATCG 3'
C) 5' TGCTAGC 3'
D) 5' CGATCGT 3'
A fragment of DNA is cloned into a plasmid with a sequencing primer-binding site. After dideoxy sequencing, the gel pattern shown in this diagram is obtained.

What was the sequence of the DNA strand that acted as the template in the sequencing reaction?
A) 5' GCTAGCA 3'
B) 5' ACGATCG 3'
C) 5' TGCTAGC 3'
D) 5' CGATCGT 3'

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
55
Use the following to answer questions
You identify an RFLP in mice by digesting genomic DNA with HindIII enzyme and radiolabeling a piece of probe DNA. Southern blot analysis shows that the probe detects a 2-kb fragment in one strain of mice and a 4-kb fragment in another strain of mice. The two mice strains are crossed to produce an F1 generation. Two F1 siblings are mated to produce a dozen F2 progeny. You isolate genomic DNA from several F1 and F2 individuals, digest with HindIII, and perform a Southern blot with the same probe.
How many bands would be seen in the Southern blot of an F2 individual?
A) one
B) two
C) three
D) four
E) one or two
You identify an RFLP in mice by digesting genomic DNA with HindIII enzyme and radiolabeling a piece of probe DNA. Southern blot analysis shows that the probe detects a 2-kb fragment in one strain of mice and a 4-kb fragment in another strain of mice. The two mice strains are crossed to produce an F1 generation. Two F1 siblings are mated to produce a dozen F2 progeny. You isolate genomic DNA from several F1 and F2 individuals, digest with HindIII, and perform a Southern blot with the same probe.
How many bands would be seen in the Southern blot of an F2 individual?
A) one
B) two
C) three
D) four
E) one or two

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
56
A pedigree and Southern blot results in humans are shown in the following figure. Filled-in figures represent individuals expressing a dominant trait (hypothetical) for blue tongue. What can you say about the potential linkage relationship between the allele responsible for the trait? Shaded regions within the homologs represent sequences that hybridize to the probe used for the Southern analysis. Arrows indicate cleavage sites used for the Southern analysis. 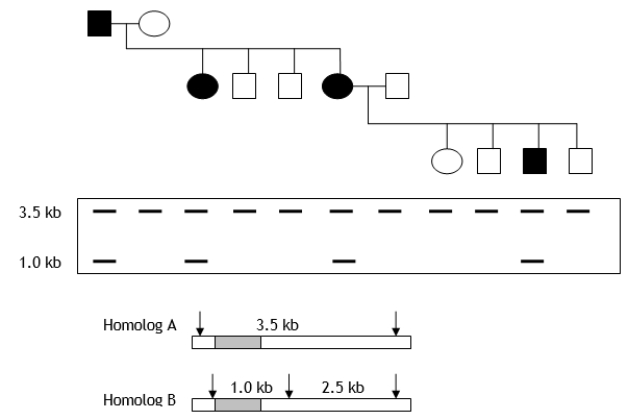


Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
57
You are trying to determine whether a certain RFLP marker is linked to a specific disease gene in dogs. A cross between a diseased male and a healthy female produces nine offspring. You isolate genomic DNA from parents and offspring, digest with EcoRI, radiolabel a portion of RFLP marker to use as a probe, and perform a Southern analysis. The pedigree and autoradiogram results are shown in the following figure. Assume that the disease is autosomal dominant. 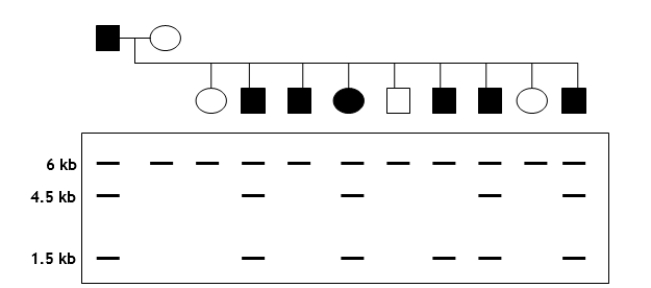 (a) Explain the relationship between the crossing results (i.e., diseased versus healthy) and the RFLP patterns.
(a) Explain the relationship between the crossing results (i.e., diseased versus healthy) and the RFLP patterns.
 (a) Explain the relationship between the crossing results (i.e., diseased versus healthy) and the RFLP patterns.
(a) Explain the relationship between the crossing results (i.e., diseased versus healthy) and the RFLP patterns.
Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
58
Cloned eukaryotic genes are not always able to be expressed in bacterial cells. What are some possible reasons for this?

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
59
What is the function of dideoxynucleotides in Sanger DNA sequencing?
A) They act as primers for DNA polymerase.
B) They act as primers for reverse transcriptase.
C) They cut the sequenced DNA at specific sites.
D) They allow only the specific sequencing of the RNAs of a genome.
E) They stop synthesis at a specific site, so the base at that site can be determined.
A) They act as primers for DNA polymerase.
B) They act as primers for reverse transcriptase.
C) They cut the sequenced DNA at specific sites.
D) They allow only the specific sequencing of the RNAs of a genome.
E) They stop synthesis at a specific site, so the base at that site can be determined.

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
60
Which of the following is NOT used in an in situ hybridization experiment?
A) chromosome spread
B) probe
C) denaturation solution
D) antibody
E) microscope
A) chromosome spread
B) probe
C) denaturation solution
D) antibody
E) microscope

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
61
Cronin et al. (Science, 2009, 325: 340-343) used RNA interference to study the immune response of the fruit fly, Drosophila melanogaster, to the bacterial pathogen Serratia marcescens. They identified several members of the JAK-STAT cell-cell signaling pathway in their analysis.
a) The PIAS gene is a negative regulator of JAK-STAT signaling (that is, it inhibits the signaling pathway). RNAi to block PIAS activity causes flies to die significantly earlier than control flies when exposed to bacteria. Does JAK-STAT signaling appear to protect the flies from the bacteria? Explain.
b) The upd gene encodes a ligand that activates JAK-STAT signaling. RNAi that blocks upd function causes flies to survive longer than control flies when exposed to bacteria. Explain how this is or is not consistent with your answer from part (a).
a) The PIAS gene is a negative regulator of JAK-STAT signaling (that is, it inhibits the signaling pathway). RNAi to block PIAS activity causes flies to die significantly earlier than control flies when exposed to bacteria. Does JAK-STAT signaling appear to protect the flies from the bacteria? Explain.
b) The upd gene encodes a ligand that activates JAK-STAT signaling. RNAi that blocks upd function causes flies to survive longer than control flies when exposed to bacteria. Explain how this is or is not consistent with your answer from part (a).

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
62
A gene in the fruit fly D. melanogaster is found to affect neuronal function when mutated. A homologue of this gene is identified in mice. Which of the following methods would it make most sense for a scientist to use to study the function of the gene in mice? (Select all that apply.)
A) Forward genetics
B) Reverse genetics
C) Mutagenesis with radiation
D) CRISPR-Cas9 genome editing targeting the gene of interest followed by nonhomologous end joining (NHEJ)
E) CRISPR-Cas9 genome editing targeting the gene of interest followed by homologous recombination (HR) with a donor template
A) Forward genetics
B) Reverse genetics
C) Mutagenesis with radiation
D) CRISPR-Cas9 genome editing targeting the gene of interest followed by nonhomologous end joining (NHEJ)
E) CRISPR-Cas9 genome editing targeting the gene of interest followed by homologous recombination (HR) with a donor template

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
63
List two applications of recombinant DNA technology.

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
64
Use the following to answer questions
A fragment of DNA is cloned into a plasmid with a sequencing primer-binding site. After dideoxy sequencing, the gel pattern shown in this diagram is obtained.
On the strand that acted as the template in the sequencing reaction, what base of the cloned fragment was closest to the primer?
A) G
B) A
C) C
D) T
A fragment of DNA is cloned into a plasmid with a sequencing primer-binding site. After dideoxy sequencing, the gel pattern shown in this diagram is obtained.

On the strand that acted as the template in the sequencing reaction, what base of the cloned fragment was closest to the primer?
A) G
B) A
C) C
D) T

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
65
Which of the following BEST describes knockout mice?
A) They have a gene of interest that has been fully disabled.
B) They have lower expression levels of a gene of interest.
C) They have higher expression levels of a gene of interest.
D) They have a point mutation in the gene of interest.
E) They have a gene removed, which results in lowered fertility.
A) They have a gene of interest that has been fully disabled.
B) They have lower expression levels of a gene of interest.
C) They have higher expression levels of a gene of interest.
D) They have a point mutation in the gene of interest.
E) They have a gene removed, which results in lowered fertility.

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
66
You try to generate a knockout mouse for a gene of interest, using the protocol described in this chapter. You do obtain progeny heterozygous for the knockout, as assessed by Southern blot. When you cross these heterozygous mice, you obtain viable progeny - some are heterozygous for the knockout, but none are found that are homozygous for the knockout. Which of following is the MOST likely explanation for this result?
A) The target gene was not disrupted successfully.
B) Genes in addition to the target gene were disrupted.
C) The heterozygous mice in your cross did not mate properly.
D) The target gene is haploinsufficient; that is, a mutant phenotype is observed when only one copy of the gene is disrupted.
E) The target gene is essential for viability.
A) The target gene was not disrupted successfully.
B) Genes in addition to the target gene were disrupted.
C) The heterozygous mice in your cross did not mate properly.
D) The target gene is haploinsufficient; that is, a mutant phenotype is observed when only one copy of the gene is disrupted.
E) The target gene is essential for viability.

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
67
Which of the following is a current concern of genetic testing? (Select all that apply.)
A) testing for a disease for which there is no cure or treatment
B) the confidentiality of the tests
C) the accuracy of the tests
D) false negative results
A) testing for a disease for which there is no cure or treatment
B) the confidentiality of the tests
C) the accuracy of the tests
D) false negative results

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
68
How might RNAi be used to treat diseases in humans?
A) siRNAs could be generated that would silence elevated levels of a harmful gene.
B) An RNAi library could be generated to assay harmful gene expression levels.
C) RNAi probes could bind to good genes and thus increase their expression.
D) RNAi could be used to amplify large amounts of therapeutic proteins.
E) RNAi cannot be used in humans, as it is a process found only in plants.
A) siRNAs could be generated that would silence elevated levels of a harmful gene.
B) An RNAi library could be generated to assay harmful gene expression levels.
C) RNAi probes could bind to good genes and thus increase their expression.
D) RNAi could be used to amplify large amounts of therapeutic proteins.
E) RNAi cannot be used in humans, as it is a process found only in plants.

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
69
A scientist mutates a gene in yeast and then looks to see what effect introducing this mutation has on the phenotype in yeast. This is an example of:
A) DNA fingerprinting.
B) next-generation sequencing.
C) transgenic research.
D) reverse genetics.
E) positional cloning.
A) DNA fingerprinting.
B) next-generation sequencing.
C) transgenic research.
D) reverse genetics.
E) positional cloning.

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
70
A scientist edits a mammalian gene by changing a single AT base pair to a GC base pair. Which of the following methods might she have used to make this change? (Select all that apply.)
A) Site-directed mutagenesis
B) Oligonucleotide-directed mutagenesis
C) CRISPR-Cas9 genome editing followed by nonhomologous end joining (NHEJ)
D) CRISPR-Cas9 genome editing followed by homologous recombination (HR) with a donor template
E) Mutagenesis with radiation
A) Site-directed mutagenesis
B) Oligonucleotide-directed mutagenesis
C) CRISPR-Cas9 genome editing followed by nonhomologous end joining (NHEJ)
D) CRISPR-Cas9 genome editing followed by homologous recombination (HR) with a donor template
E) Mutagenesis with radiation

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
71
Which of the following remains a problem of gene therapy?
A) To date, there has not been a case of gene therapy curing a disease.
B) We have yet to develop a vector for delivering the gene.
C) Patients mount immune responses to the transferred gene and vectors.
D) We do not have the ability to use somatic gene therapy at this time.
E) We can only alter germ-line cells at this time.
A) To date, there has not been a case of gene therapy curing a disease.
B) We have yet to develop a vector for delivering the gene.
C) Patients mount immune responses to the transferred gene and vectors.
D) We do not have the ability to use somatic gene therapy at this time.
E) We can only alter germ-line cells at this time.

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck
72
The following are steps required to develop a knockout mouse. Place them in the CORRECT order.
A) 1, 4, 5, 6, 2, 3
B) 4, 1, 6, 5, 3, 2
C) 4, 5, 1, 6, 2, 3
D) 1, 4, 5, 2, 6, 3
E) 1, 6, 4, 5, 2, 3
A) 1, 4, 5, 6, 2, 3
B) 4, 1, 6, 5, 3, 2
C) 4, 5, 1, 6, 2, 3
D) 1, 4, 5, 2, 6, 3
E) 1, 6, 4, 5, 2, 3

Unlock Deck
Unlock for access to all 72 flashcards in this deck.
Unlock Deck
k this deck


