Deck 17: Control of Gene Expression in Eukaryotes
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/64
Play
Full screen (f)
Deck 17: Control of Gene Expression in Eukaryotes
1
In Arabidopsis, FLD (a deacetylase enzyme) stimulates flowering. Which of the following statements is TRUE?
A) FLD deacetylates histones that bind to regions of the FLC gene and stimulates its transcription.
B) FLD deacetylates histones surrounding the FLD gene, causing suppression of FLD transcription.
C) FLD deacetylates histones that bind to the FLC gene, causing repression of FLC transcription.
D) FLD causes repression of FLC translation.
A) FLD deacetylates histones that bind to regions of the FLC gene and stimulates its transcription.
B) FLD deacetylates histones surrounding the FLD gene, causing suppression of FLD transcription.
C) FLD deacetylates histones that bind to the FLC gene, causing repression of FLC transcription.
D) FLD causes repression of FLC translation.
C
2
Proteins that affect the structure of DNA bound to histones without altering histone chemical structure are called:
A) RISCs.
B) chromatin-remodeling complexes.
C) splicing complexes.
D) MRE activator proteins.
E) dicers.
A) RISCs.
B) chromatin-remodeling complexes.
C) splicing complexes.
D) MRE activator proteins.
E) dicers.
B
3
In both prokaryotes and eukaryotes, groups of genes can be regulated simultaneously (coordinately expressed). However, each group accomplishes this task differently. Explain how coordinate expression differs in prokaryotes compared with eukaryotes.
not answered
4
A scientist has discovered a new gene that she believes encodes a transcriptional activator protein. She is trying to determine where this protein binds. Which of the following describes a technique she could use to answer her question?
A) Chromatin immunoprecipitation to determine the DNA sequences to which the putative transcriptional activator protein binds
B) RNA interference to determine the RNA sequences to which the putative transcriptional activator protein binds
C) Crosslinking to determine the interactions the putative transcriptional activator protein has with other proteins
D) Chromative immunoprecipitation to determine the proteins to which the putative transcriptional activator protein binds
E) RNA interference combined with crosslinking to determine all DNA and RNA sequences to which the putative transcriptional activator protein binds
A) Chromatin immunoprecipitation to determine the DNA sequences to which the putative transcriptional activator protein binds
B) RNA interference to determine the RNA sequences to which the putative transcriptional activator protein binds
C) Crosslinking to determine the interactions the putative transcriptional activator protein has with other proteins
D) Chromative immunoprecipitation to determine the proteins to which the putative transcriptional activator protein binds
E) RNA interference combined with crosslinking to determine all DNA and RNA sequences to which the putative transcriptional activator protein binds

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
5
Which of the following events is known to make DNA more sensitive to digestion by DNase I?
A) DNA methylation
B) deacetylation of histone
C) acetylation of histone
D) formation of heterochromatin
A) DNA methylation
B) deacetylation of histone
C) acetylation of histone
D) formation of heterochromatin

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
6
What is the consequence of methylation of DNA sequences called CpG islands?
A) active transcription
B) transcription repression
C) recruitment of chromosome-remodeling complex
D) transcriptional stalling
E) insulator sequence formation
A) active transcription
B) transcription repression
C) recruitment of chromosome-remodeling complex
D) transcriptional stalling
E) insulator sequence formation

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
7
What is the connection between DNA methylation, histone deacetylation, and gene regulation in eukaryotes?

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
8
When regions around genes become sensitive to the enzyme DNase I, this is an indication that those regions are:
A) becoming transcriptionally active.
B) becoming more condensed.
C) binding to the single-strand binding proteins.
D) destabilizing and transcriptionally inactive.
E) becoming highly methylated by a methylase.
A) becoming transcriptionally active.
B) becoming more condensed.
C) binding to the single-strand binding proteins.
D) destabilizing and transcriptionally inactive.
E) becoming highly methylated by a methylase.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
9
Which of the following molecules is capable of targeting chromatin-remodeling complexes to specific DNA sequences to modify chromatin structure and activate gene expression?
A) transcriptional repressor
B) enhancer
C) DNAse I
D) transcriptional activator
E) histone deacetylase
A) transcriptional repressor
B) enhancer
C) DNAse I
D) transcriptional activator
E) histone deacetylase

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
10
What would be the advantage of regulating gene expression at many levels rather than simply regulating at one level (such as at the start of transcription)?

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
11
If a deacetylase inhibitor were injected into a mouse brain, which of the following would you expect to occur?
A) A general increase in gene expression would occur.
B) A general decrease in gene expression would occur.
C) A general increase or decrease in gene expression, depending on the gene, would occur.
D) Acetyl groups would be removed from histones.
E) Methyl groups would be removed from the DNA.
A) A general increase in gene expression would occur.
B) A general decrease in gene expression would occur.
C) A general increase or decrease in gene expression, depending on the gene, would occur.
D) Acetyl groups would be removed from histones.
E) Methyl groups would be removed from the DNA.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
12
List at least two ways that regulation of genes for yeast galactose-metabolizing enzymes is different from regulation of E. coli lactose-utilizing enzymes.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
13
Which of the following statements about regulation of eukaryotic gene expression is INCORRECT?
A) The presence of a nuclear membrane separating transcription and translation in eukaryotes led to the evolution of additional mechanisms of gene regulation.
B) In eukaryotes, most structural genes are found within operons.
C) Eukaryotic mRNAs are generally more stable than prokaryotic mRNAs.
D) The rate of degradation of mRNAs is important in regulation in eukaryotes.
E) Posttranslational regulation of histones is unique to eukaryotes.
A) The presence of a nuclear membrane separating transcription and translation in eukaryotes led to the evolution of additional mechanisms of gene regulation.
B) In eukaryotes, most structural genes are found within operons.
C) Eukaryotic mRNAs are generally more stable than prokaryotic mRNAs.
D) The rate of degradation of mRNAs is important in regulation in eukaryotes.
E) Posttranslational regulation of histones is unique to eukaryotes.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
14
Which of the following statements about CpG islands is CORRECT?
A) CpG islands are commonly found at the 3' UTR regions.
B) The CpG island methylation is universal across both prokaryotes and eukaryotes.
C) Methylated CpG islands are associated with long-term gene repression.
D) Transcriptionally active DNA has a higher frequency of methylated CpG.
E) There is an association between DNA methylation at the CpG island and acetylation of histone via recruitment of acetylases.
A) CpG islands are commonly found at the 3' UTR regions.
B) The CpG island methylation is universal across both prokaryotes and eukaryotes.
C) Methylated CpG islands are associated with long-term gene repression.
D) Transcriptionally active DNA has a higher frequency of methylated CpG.
E) There is an association between DNA methylation at the CpG island and acetylation of histone via recruitment of acetylases.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
15
A scientist is using crosslinked chromatin immunoprecipitation (XChIP) to determine the DNA sequences to which a protein of interest binds. However, when she attempts to sequence the DNA in the last step of the procedure, she is unable to obtain any DNA to sequence. Which of the following is a likely reason she is getting these results?
A) The antibody she is using to precipitate the protein-DNA complex is not specific enough and binds the protein in question and five other proteins.
B) The protein and DNA are not properly crosslinked.
C) The antibody she is using is not specific to the DNA sequence to which the protein binds.
D) The DNA is too tightly complexed with histone proteins to allow sequencing.
E) The cells were lysed during the precipitation stage of the process.
A) The antibody she is using to precipitate the protein-DNA complex is not specific enough and binds the protein in question and five other proteins.
B) The protein and DNA are not properly crosslinked.
C) The antibody she is using is not specific to the DNA sequence to which the protein binds.
D) The DNA is too tightly complexed with histone proteins to allow sequencing.
E) The cells were lysed during the precipitation stage of the process.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
16
Choose the CORRECT order of procedure in performing chromatin immunoprecipitation (ChIP). 1. DNA sequencing and fragment identification
2) Removal of crosslinking and separation of DNA and proteins
3) Crosslinking proteins and chromosomes via UV
4) Antibody incubation for immunoprecipitation
5) Protein degradation via proteases
6) Cell lysis and chromosome fragmentation
A) 1, 3, 2, 4, 5, 6
B) 2, 6, 5, 1, 3, 4
C) 4, 3, 2, 5, 1, 5
D) 3, 6, 4, 2, 5, 1
E) 5, 1, 3, 6, 4, 2
2) Removal of crosslinking and separation of DNA and proteins
3) Crosslinking proteins and chromosomes via UV
4) Antibody incubation for immunoprecipitation
5) Protein degradation via proteases
6) Cell lysis and chromosome fragmentation
A) 1, 3, 2, 4, 5, 6
B) 2, 6, 5, 1, 3, 4
C) 4, 3, 2, 5, 1, 5
D) 3, 6, 4, 2, 5, 1
E) 5, 1, 3, 6, 4, 2

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
17
Which of the following CANNOT be determined by chromatin immunoprecipitation (ChIP)?
A) the types of modification present on the DNA-binding proteins
B) the location of modified histones that activate or repress transcription
C) the position of transcription factors and associated regulators on the chromosome
D) identification of active promoters on a genome-wide level
E) the exact amount of protein expressed from a gene
A) the types of modification present on the DNA-binding proteins
B) the location of modified histones that activate or repress transcription
C) the position of transcription factors and associated regulators on the chromosome
D) identification of active promoters on a genome-wide level
E) the exact amount of protein expressed from a gene

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
18
Histone methylation can have many different effects on gene expression. In some cases, histone methylation is associated with activation of transcription, whereas in other cases it can trigger the formation of heterochromatin and a decrease in transcription. If histone methylation has been detected in the region of gene X in yeast, describe an experiment that could distinguish whether the methylation is important to activate or repress transcription of gene X.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
19
Which of the following statements about histones and gene expression is CORRECT?
A) In a general sense, highly condensed DNA bound with histone proteins represses gene expression.
B) Acetylation involves the addition of acetyl groups to histone proteins, and it usually results in repression of transcription.
C) Addition of methyl groups to the tails of histone proteins always results in activation of transcription.
D) Histone code refers to the modification that takes place on the globular domain of the octamer histone core.
E) Phosphorylation of cytosines generally leads to increase in transcription..
A) In a general sense, highly condensed DNA bound with histone proteins represses gene expression.
B) Acetylation involves the addition of acetyl groups to histone proteins, and it usually results in repression of transcription.
C) Addition of methyl groups to the tails of histone proteins always results in activation of transcription.
D) Histone code refers to the modification that takes place on the globular domain of the octamer histone core.
E) Phosphorylation of cytosines generally leads to increase in transcription..

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
20
What are three ways in which gene regulation is accomplished by modifying the structure of chromatin?

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
21
Which of the following statements about response elements is INCORRECT?
A) A single eukaryotic gene is regulated by only one unique response element.
B) Response elements are composed of specific consensus sequences that are unique from one another.
C) Different genes can possess a common regulatory element upstream of their start site.
D) Multiple response elements allow the same gene to be activated by different stimuli.
E) Response elements allow complex biochemical responses in eukaryotic cells.
A) A single eukaryotic gene is regulated by only one unique response element.
B) Response elements are composed of specific consensus sequences that are unique from one another.
C) Different genes can possess a common regulatory element upstream of their start site.
D) Multiple response elements allow the same gene to be activated by different stimuli.
E) Response elements allow complex biochemical responses in eukaryotic cells.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
22
Consider the regulation of galactose metabolism through GAL4. Which of the following would result from a mutation that allowed GAL3 to bind to GAL80 in the absence of galactose?
A) Transcription of the genes involved in galactose metabolism would occur both in the presence and in the absence of galactose.
B) GAL80 would be able to bind to GAL4, and transcription of the genes involved in galactose metabolism would be repressed.
C) GAL4 would no longer be able to bind to the DNA; thus, transcription of the genes involved in galactose metabolism would occur.
D) GAL80 would no longer be able to stimulate transcription of the genes involved in galactose metabolism.
E) There would be no change in the regulation of galactose metabolism because GAL3 normally binds to GAL80 to cause a conformation change in GAL80.
A) Transcription of the genes involved in galactose metabolism would occur both in the presence and in the absence of galactose.
B) GAL80 would be able to bind to GAL4, and transcription of the genes involved in galactose metabolism would be repressed.
C) GAL4 would no longer be able to bind to the DNA; thus, transcription of the genes involved in galactose metabolism would occur.
D) GAL80 would no longer be able to stimulate transcription of the genes involved in galactose metabolism.
E) There would be no change in the regulation of galactose metabolism because GAL3 normally binds to GAL80 to cause a conformation change in GAL80.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
23
Although operons are not common in eukaryotes, eukaryotic genes may be activated by the same stimulus. Which of the following DNA regulatory sequences makes this coordinated gene expression possible?
A) core promoter
B) enhancer element
C) response element
D) boundary element
E) silencer element
A) core promoter
B) enhancer element
C) response element
D) boundary element
E) silencer element

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
24
We learned in Chapter 17 that GAL3 protein binds to GAL80 and frees up GAL4 to bind to UAS and activate transcription of genes involved in galactose metabolism. Assuming that you did not have prior knowledge of GAL3 protein but knew about GAL4 and GAL80, what experiments would you perform to identify the GAL3 gene and additional genes involved in the regulation of GAL4?

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
25
Which of the following sequences or molecules is LEAST relevant to the assembly of the basal transcription apparatus for transcription?
A) core promoter
B) general transcription factors
C) TATA box
D) RNA polymerase
E) enhancer
A) core promoter
B) general transcription factors
C) TATA box
D) RNA polymerase
E) enhancer

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
26
What type of cellular activity is known to occur within P bodies?
A) active transcription
B) active translation
C) RNA degradation
D) transcriptional stalling
E) DNA hypermethylation
A) active transcription
B) active translation
C) RNA degradation
D) transcriptional stalling
E) DNA hypermethylation

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
27
Which of the following eukaryotic gene regulatory mechanisms acts after transcription has been initiated?
A) changes in chromatin structure
B) changes in transcriptional regulator proteins
C) assembly of the basal transcription apparatus
D) factors that increase RNA polymerase stalling
E) association of transcriptional coactivators
A) changes in chromatin structure
B) changes in transcriptional regulator proteins
C) assembly of the basal transcription apparatus
D) factors that increase RNA polymerase stalling
E) association of transcriptional coactivators

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
28
Given the following figure, what would be the effect of a mutation that occurred in the insulator that prevented the binding of the insulator-binding protein? 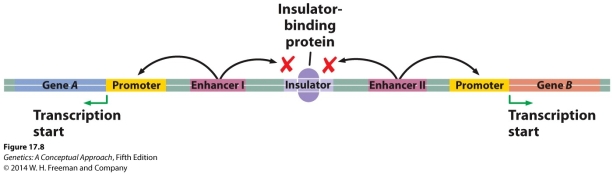
A) Both Enhancer 1 and Enhancer II would be able to stimulate the transcription of Genes A and B.
B) Enhancer I would no longer be able to stimulate the transcription of Gene B.
C) Enhancer I will be able to hypermethylate Gene B.
D) As long as the insulator sequence is not deleted, there will be no change in the transcription regulation of Gene A or B.
E) Enhancer II will no longer be able to stimulate the transcription of Gene A.

A) Both Enhancer 1 and Enhancer II would be able to stimulate the transcription of Genes A and B.
B) Enhancer I would no longer be able to stimulate the transcription of Gene B.
C) Enhancer I will be able to hypermethylate Gene B.
D) As long as the insulator sequence is not deleted, there will be no change in the transcription regulation of Gene A or B.
E) Enhancer II will no longer be able to stimulate the transcription of Gene A.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
29
mRNAs are degraded by enzymes called:
A) DNAse I.
B) ribozymes.
C) heat-shock proteins.
D) silencers.
E) ribonucleases.
A) DNAse I.
B) ribozymes.
C) heat-shock proteins.
D) silencers.
E) ribonucleases.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
30
Degradation of a eukaryotic mRNA is generally initiated by:
A) cleavage of the 5' end.
B) random cleavages throughout the mRNA strand.
C) shortening of the poly(A) tail.
D) recruitment of the chromosome-remodeling complex.
E) removal of the 5' cap.
A) cleavage of the 5' end.
B) random cleavages throughout the mRNA strand.
C) shortening of the poly(A) tail.
D) recruitment of the chromosome-remodeling complex.
E) removal of the 5' cap.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
31
Which of the following facts about eukaryotic gene regulation is TRUE?
A) Transcriptional activator proteins bind to the DNA in a nonspecific manner.
B) Eukaryotic enhancers are a part of the basal transcription apparatus.
C) The eukaryotic regulatory promoters are highly conserved with the same consensus sequences throughout the genome.
D) Mediators are protein complexes involved in regulating transcription rates.
E) The transcriptional repressors always bind to the insulator elements.
A) Transcriptional activator proteins bind to the DNA in a nonspecific manner.
B) Eukaryotic enhancers are a part of the basal transcription apparatus.
C) The eukaryotic regulatory promoters are highly conserved with the same consensus sequences throughout the genome.
D) Mediators are protein complexes involved in regulating transcription rates.
E) The transcriptional repressors always bind to the insulator elements.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
32
A eukaryotic DNA sequence that affects transcription at distant promoters is called a(n):
A) insulator.
B) silencer.
C) mediator.
D) enhancer.
E) repressor.
A) insulator.
B) silencer.
C) mediator.
D) enhancer.
E) repressor.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
33
Insulators can block the effects of enhancers only when they lie:
A) between an enhancer and a promoter.
B) upstream of a promoter.
C) adjacent to a promoter.
D) within the structural genes.
E) within a consensus sequence.
A) between an enhancer and a promoter.
B) upstream of a promoter.
C) adjacent to a promoter.
D) within the structural genes.
E) within a consensus sequence.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
34
Eukaryotic genes can be introduced into bacteria by recombinant DNA techniques. If the introduced gene encodes a protein that is also found in bacteria-for example, a universally used glycolysis enzyme-then expression of the eukaryotic gene may produce a protein that functions in the bacterial cell.
The mouse gene for a glycolysis enzyme is introduced into an E. coli cell that has a mutant gene for the bacterial version of the same enzyme. Even though the mouse enzyme should function in the bacterial cell and restore the cell's ability to perform glycolysis, it does not.
Provide two possible reasons why this experiment does not work and propose a solution to overcome one of the problems you suggest.
The mouse gene for a glycolysis enzyme is introduced into an E. coli cell that has a mutant gene for the bacterial version of the same enzyme. Even though the mouse enzyme should function in the bacterial cell and restore the cell's ability to perform glycolysis, it does not.
Provide two possible reasons why this experiment does not work and propose a solution to overcome one of the problems you suggest.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
35
A boundary element is also known as a(n):
A) insulator.
B) repressor.
C) enhancer.
D) coactivator.
E) mediator.
A) insulator.
B) repressor.
C) enhancer.
D) coactivator.
E) mediator.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
36
What are two distinct functions that transcriptional activator proteins perform in order to regulate gene transcription?

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
37
DNA sequences that might act as enhancers (regulatory elements) can be attached to the gene for green fluorescent protein (GFP) and the combined DNA sequence can be reintroduced into an organism. If the DNA sequence attached to GFP actually can act as an enhancer, glowing green pigment will be observed in particular regions of the organism. Gene A is expressed in developing nerve cells, muscle cells, and intestinal cells in the fruit fly embryo. The region of DNA around gene A is depicted below. The pieces of DNA indicated with lines 1-5 were attached to GFP and tested for the ability to activate gene expression in the fly embryo. In piece 5, the "X" indicates a deletion of a single base pair. 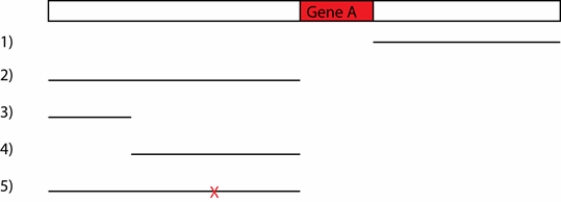
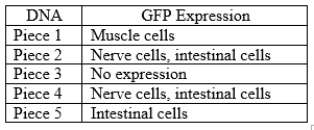 a. Based on the pattern of GFP expression when attached to either piece 1 or piece 2, propose a model for how gene A is activated in the different cells of the embryo.
a. Based on the pattern of GFP expression when attached to either piece 1 or piece 2, propose a model for how gene A is activated in the different cells of the embryo.
b. Propose an explanation for the difference in GFP expression caused by piece 3 and piece 4.
c. What has the deletion "X" in piece 5 likely affected to cause the change of GFP expression compared to piece 2? How does this result refine the model proposed in (a)?

 a. Based on the pattern of GFP expression when attached to either piece 1 or piece 2, propose a model for how gene A is activated in the different cells of the embryo.
a. Based on the pattern of GFP expression when attached to either piece 1 or piece 2, propose a model for how gene A is activated in the different cells of the embryo.b. Propose an explanation for the difference in GFP expression caused by piece 3 and piece 4.
c. What has the deletion "X" in piece 5 likely affected to cause the change of GFP expression compared to piece 2? How does this result refine the model proposed in (a)?

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
38
Which type of control is illustrated by GAL4 in the control of genes for yeast galactose-metabolizing enzymes?
A) negative inducible
B) negative repressible
C) positive inducible
D) positive repressible
A) negative inducible
B) negative repressible
C) positive inducible
D) positive repressible

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
39
A mutation in the gene for the yeast regulatory protein GAL4 causes yeast to grow poorly on galactose. What is the function of GAL4?
A) It is a substrate that binds and activates a transcriptional activator.
B) It is a product that binds and activates a transcriptional repressor.
C) It is a transcriptional activator for the galactose-digesting enzyme gene.
D) It is a transcriptional repressor that prevents expression of yeast galactose-digesting enzymes.
E) It is an enzyme that metabolizes galactose.
A) It is a substrate that binds and activates a transcriptional activator.
B) It is a product that binds and activates a transcriptional repressor.
C) It is a transcriptional activator for the galactose-digesting enzyme gene.
D) It is a transcriptional repressor that prevents expression of yeast galactose-digesting enzymes.
E) It is an enzyme that metabolizes galactose.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
40
The SUC2 gene in yeast encodes an enzyme to convert the sugar, sucrose, into glucose and fructose, which is necessary for yeast to use sucrose as a source of food. In the presence of glucose, SUC2 expression is switched off. But in the absence of glucose, SUC2 expression increases 100-fold. The expression of SUC2 in wild-type yeast and two mutant yeast strains is shown below. The mutations occur in two genes other than SUC2: SNF1 and SSN6. a. Is the SNF1 gene normally important for activation or repression of SUC2 expression?
b. Is the SSN6 gene normally important for activation or repression of SUC2 expression?
c. If SNF1 and SSN6 work in the same pathway to regulate SUC2 expression, the order in which the genes acts can be one of two possibilities (where indicates activation and indicates inhibition): (1) SNF1 SSN6 SUC2
SSN6 acts to repress SUC2 expression, and SNF1 activates SUC2 by inhibiting the inhibitor, SSN6.
(2) SSN6 SNF1 SUC2
SNF1 activates expression, and SSN6 represses by inhibiting the action of SNF1. Design an experiment to distinguish between these two possible orders. What would you expect the outcome of your experiment to be in each of the two possible cases?
b. Is the SSN6 gene normally important for activation or repression of SUC2 expression?
c. If SNF1 and SSN6 work in the same pathway to regulate SUC2 expression, the order in which the genes acts can be one of two possibilities (where indicates activation and indicates inhibition): (1) SNF1 SSN6 SUC2
SSN6 acts to repress SUC2 expression, and SNF1 activates SUC2 by inhibiting the inhibitor, SSN6.
(2) SSN6 SNF1 SUC2
SNF1 activates expression, and SSN6 represses by inhibiting the action of SNF1. Design an experiment to distinguish between these two possible orders. What would you expect the outcome of your experiment to be in each of the two possible cases?

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
41
What are four things that influence the stability of eukaryotic mRNAs?

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
42
In the nematode roundworm Caenorhabditis elegans, the LIN-14 protein controls the timing of certain cell divisions during development. LIN-14 protein levels are normally high in early development but decrease in the later stages. In a lin-4 mutant, the level of LIN-14 protein stays high throughout development, changing the pattern of cell divisions in the animal and producing defects in the shape of the animal. The lin-4 gene encodes a microRNA that binds to a sequence in the 3'UTR of the lin-14 mRNA.
a. How does the lin-4 microRNA likely regulate LIN-14 protein levels? Explain why the lin-4 mutant has high levels of LIN-14 throughout development.
b. Mutations in the 3' UTR of lin-14 have been identified that alter the sequence to which lin-4 normally binds. What effect would these mutations be expected to have on the expression of LIN-14 protein in the animal?
a. How does the lin-4 microRNA likely regulate LIN-14 protein levels? Explain why the lin-4 mutant has high levels of LIN-14 throughout development.
b. Mutations in the 3' UTR of lin-14 have been identified that alter the sequence to which lin-4 normally binds. What effect would these mutations be expected to have on the expression of LIN-14 protein in the animal?

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
43
Over the past decade, a significant finding in biology has been the identification of miRNAs and siRNAs and their role in regulating the development of many multicellular organisms. Briefly describe the four different ways these small RNAs influence gene expression.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
44
Consider the way alternative splicing leads to the production of the small t antigen and the large T antigen in the mammalian virus SV40. What would be the result of a mutation that occurred in the gene that encodes the SF2 protein?
A) Higher amounts of the small t antigen would be produced.
B) Use of the first 5ʹ splice site would be favored over the second 5ʹ splice site.
C) The large T antigen would be produced but at lower than normal levels.
D) Both the first and second 5ʹ splice sites could be used, but the second would be favored over the first.
E) Regions A, B, and C would all be transcribed and translated to produce the correct antigen.
A) Higher amounts of the small t antigen would be produced.
B) Use of the first 5ʹ splice site would be favored over the second 5ʹ splice site.
C) The large T antigen would be produced but at lower than normal levels.
D) Both the first and second 5ʹ splice sites could be used, but the second would be favored over the first.
E) Regions A, B, and C would all be transcribed and translated to produce the correct antigen.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
45
Which of the following mechanisms does NOT involve siRNA- and miRNA-based gene regulation?
A) cleavage of mRNA
B) inhibition of translation
C) posttranslational modification
D) degradation of mRNA
E) transcriptional silencing
A) cleavage of mRNA
B) inhibition of translation
C) posttranslational modification
D) degradation of mRNA
E) transcriptional silencing

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
46
Regulation of gene expression using siRNAs is found in:
A) prokaryotes only.
B) eukaryotes only.
C) prokaryotes and eukaryotes.
A) prokaryotes only.
B) eukaryotes only.
C) prokaryotes and eukaryotes.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
47
RITS consists of:
A) siRNAs and proteins.
B) miRNAs and proteins.
C) RISCs and mRNAs.
D) methyl groups and histone proteins.
E) DNA, histone proteins, and mRNAs.
A) siRNAs and proteins.
B) miRNAs and proteins.
C) RISCs and mRNAs.
D) methyl groups and histone proteins.
E) DNA, histone proteins, and mRNAs.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
48
Suppose a scientist discovered a mutant strain of C. elegans worms that had lost the ability to regulate gene expression via RNA interference mechanisms. Which of the following might she predict is a possible source of this defect?
A) There is a mutation in the gene that encodes the Dicer enzyme that makes it nonfunctional.
B) There is a mutation that results in an overexpression of siRNAs.
C) There is a mutation in the gene that encodes the Slicer enzyme that allows it to cut RNA more efficiently.
D) There is a mutation in a gene that encodes an acetylase enzyme that prevents histone acetylation.
E) There is a mutation in a gene that encodes an enzyme that selects the correct splice site for alternative splicing.
A) There is a mutation in the gene that encodes the Dicer enzyme that makes it nonfunctional.
B) There is a mutation that results in an overexpression of siRNAs.
C) There is a mutation in the gene that encodes the Slicer enzyme that allows it to cut RNA more efficiently.
D) There is a mutation in a gene that encodes an acetylase enzyme that prevents histone acetylation.
E) There is a mutation in a gene that encodes an enzyme that selects the correct splice site for alternative splicing.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
49
Define RNA silencing (or interference). Explain how siRNAs arise and how they potentially affect gene expression. How are siRNAs different from the antisense RNA mechanism?

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
50
A fly (Drosophila) with an XY genotype has a mutation in its sex-lethal (Sxl) gene that renders its protein product nonfunctional. Which of the following describes the sex of this fly?
A) male
B) female
C) intersex
D) male, but it will be sterile
E) female, but it will be sterile
A) male
B) female
C) intersex
D) male, but it will be sterile
E) female, but it will be sterile

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
51
Describe the unusual posttranscriptional control of Drosophila sex determination. How does the cascade of events differ between males and females?

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
52
Which of the following processes is also known as RNA silencing or posttranscriptional gene silencing?
A) protein degradation
B) transcriptional stalling
C) RNA splicing
D) transcriptional repression
E) RNA interference
A) protein degradation
B) transcriptional stalling
C) RNA splicing
D) transcriptional repression
E) RNA interference

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
53
A genetics student identified a male fly (Drosophila) that had an XX genotype. Which of the following is the MOST accurate plausible explanation?
A) The fly could only have a mutation in its sex-lethal (Sxl) gene.
B) The fly could only have a mutation in its transformer (tra) gene.
C) The fly could only have a mutation in its double-sex (dsx) gene.
D) The fly could have a mutation in either its sex-lethal (Sxl) or transformer (tra) gene.
E) The fly could have a mutation in its sex-lethal (Sxl), transformer (tra), or double-sex (dsx) gene.
A) The fly could only have a mutation in its sex-lethal (Sxl) gene.
B) The fly could only have a mutation in its transformer (tra) gene.
C) The fly could only have a mutation in its double-sex (dsx) gene.
D) The fly could have a mutation in either its sex-lethal (Sxl) or transformer (tra) gene.
E) The fly could have a mutation in its sex-lethal (Sxl), transformer (tra), or double-sex (dsx) gene.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
54
Explain how the poly(A)-binding protein that binds to the poly(A) tails located in the 3' end of an mRNA can play a key role in an mRNA degradation pathway that proceeds from the 5'end of an mRNA in a 5' 3' direction.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
55
Alternative splicing is known to be important in the regulation of the:
A) production of heat-shock elements.
B) mammalian SV40 virus.
C) lac operon in E. coli.
D) metallothionein gene.
E) None of the answers is correct.
A) production of heat-shock elements.
B) mammalian SV40 virus.
C) lac operon in E. coli.
D) metallothionein gene.
E) None of the answers is correct.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
56
A scientist created transgenic C. elegans worms that expressed a gene that made the worms glow green under fluorescent light. Then she injected double-stranded RNA complementary to this gene. Which of the following results was MOST likely to be seen?
A) The worms glowed green under fluorescent light.
B) The worms were not able to glow green under fluorescent light.
C) The injection of double-stranded RNA was lethal and the worms died.
D) The worms had an increased life span.
E) The CpG islands near the promoter of the gene became demethylated.
A) The worms glowed green under fluorescent light.
B) The worms were not able to glow green under fluorescent light.
C) The injection of double-stranded RNA was lethal and the worms died.
D) The worms had an increased life span.
E) The CpG islands near the promoter of the gene became demethylated.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
57
In which part of the mRNA does degradation generally begin?
A) at the 5' end with the removal of the poly(A) tail
B) at the 5' end with the removal of the methyl cap
C) at the 3' end with the removal of the poly(A) tail
D) at the 3' end with the removal of the methyl cap
E) Removal from either end is equally likely.
A) at the 5' end with the removal of the poly(A) tail
B) at the 5' end with the removal of the methyl cap
C) at the 3' end with the removal of the poly(A) tail
D) at the 3' end with the removal of the methyl cap
E) Removal from either end is equally likely.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
58
Attenuation occurs in prokaryotes but has not been seen as a gene-expression mechanism in eukaryotes. Why not?

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
59
When siRNAs are present, the rate of mRNA degradation _____ and the rate of protein production _____.
A) increases; increases
B) increases; decreases
C) decreases; decreases
D) decreases; increases
E) stays constant; decreases
A) increases; increases
B) increases; decreases
C) decreases; decreases
D) decreases; increases
E) stays constant; decreases

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
60
siRNAs and miRNAs are produced by the:
A) cleavage of RISCs by endonucleases.
B) cleavage of functional mRNA within the cytoplasm.
C) cleavage of pre-mRNA in the nucleus.
D) cutting and processing of double-stranded RNA by Dicer enzymes.
E) cutting and processing of double-stranded RNA by Slicer enzymes.
A) cleavage of RISCs by endonucleases.
B) cleavage of functional mRNA within the cytoplasm.
C) cleavage of pre-mRNA in the nucleus.
D) cutting and processing of double-stranded RNA by Dicer enzymes.
E) cutting and processing of double-stranded RNA by Slicer enzymes.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
61
Which gene-control mechanisms from the previous question are not used in prokaryotes, and why?

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
62
List five levels at which control of gene activity can take place in eukaryotes.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
63
Let us assume that your goal is to determine whether or not the expression of your favorite gene (YFG) is regulated by miRNA. Also assume that the genome sequence is known for the organism from which YFG was isolated. Describe the logical experiment steps to achieve your goal.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck
64
After translation, eukaryotic proteins can be modified by:
A) acetylation.
B) the addition of phosphate groups.
C) the removal of amino acids.
D) the addition of methyl groups.
E) acetylation, addition of phosphate and methyl groups, or the removal of amino acids.
A) acetylation.
B) the addition of phosphate groups.
C) the removal of amino acids.
D) the addition of methyl groups.
E) acetylation, addition of phosphate and methyl groups, or the removal of amino acids.

Unlock Deck
Unlock for access to all 64 flashcards in this deck.
Unlock Deck
k this deck


