Deck 13: Amplification Techniques
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/45
Play
Full screen (f)
Deck 13: Amplification Techniques
1
Taq polymerase can add nucleotide bases to a DNA strand at a rate of about
A)10 dNTPs per minute.
B)1000 dNTPs per minute.
C)100 dNTPs per second.
D)10 dNTPs per second.
E)100 dNTPs per minute.
A)10 dNTPs per minute.
B)1000 dNTPs per minute.
C)100 dNTPs per second.
D)10 dNTPs per second.
E)100 dNTPs per minute.
100 dNTPs per second.
2
The composition of which PCR reagent below is most critical in insuring that amplification of only the target DNA occurs?
A)dNTPs
B)Template DNA extraction buffer
C)Oligonucleotide primers
D)DNA polymerase
E)PCR reaction buffer
A)dNTPs
B)Template DNA extraction buffer
C)Oligonucleotide primers
D)DNA polymerase
E)PCR reaction buffer
Oligonucleotide primers
3
Which technique below is used to amplify messenger RNA (mRNA)?
A)Branched chain PCR
B)Ligase chain reaction
C)Reverse transcriptase PCR
D)Molecular beacon PCR
E)TaqMan PCR
A)Branched chain PCR
B)Ligase chain reaction
C)Reverse transcriptase PCR
D)Molecular beacon PCR
E)TaqMan PCR
Reverse transcriptase PCR
4
Review the protocol herein and identify its name. Two labeled probes attached to target DNA. DNA polymerase fills in the nucleotides between the p and DNA ligase joins the two pieces. The product is denatured into single strands, and the process i repeated. Amplified target DNA is detected via the labels on the probes.
A)Molecular beacon PCR
B)Ligase chain reaction
C)5' RACE PCR
D)Branched chain PCR
E)Nucleic Acid Sequence Based Amplification
A)Molecular beacon PCR
B)Ligase chain reaction
C)5' RACE PCR
D)Branched chain PCR
E)Nucleic Acid Sequence Based Amplification

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
5
How is DNA denaturation (conversion to single stranded form)usually accomplished in PCR?
A)Alkaline pH
B)Formaldehyde
C)Heat
D)Any of the above, depending on the target DNA sequence
A)Alkaline pH
B)Formaldehyde
C)Heat
D)Any of the above, depending on the target DNA sequence

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
6
If the optimal pH for Taq polymerase is about 7.2, why are the reaction buffers manufactured to have a pH of about 8.5?
A)Unincorporated nucleotides are unstable at neutral to acidic pHs.
B)Hydrogen bonds form poorly at neutral to acidic pHs.
C)Optimal reaction conditions for Taq polymerase are generally not used because rapid incorporation of nucleotides into a growing DNA chains leads to more errors.
D)pH is temperature dependent, and at the optimal temperature for Taq polymerase to function, the buffer pH will decrease.
A)Unincorporated nucleotides are unstable at neutral to acidic pHs.
B)Hydrogen bonds form poorly at neutral to acidic pHs.
C)Optimal reaction conditions for Taq polymerase are generally not used because rapid incorporation of nucleotides into a growing DNA chains leads to more errors.
D)pH is temperature dependent, and at the optimal temperature for Taq polymerase to function, the buffer pH will decrease.

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
7
Why is intact mRNA so hard to purify?
A)It is inherently a molecule with a short life.
B)RNases are relatively resistant to destruction.
C)RNases are present in almost every environment, including on human skin.
D)All of the above.
A)It is inherently a molecule with a short life.
B)RNases are relatively resistant to destruction.
C)RNases are present in almost every environment, including on human skin.
D)All of the above.

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
8
Which detection method below is frequently used in real time PCR to detect double- stranded DNA product?
A)Biotin
B)SYBR Green II dye
C)Propidium iodide
D)SYBR Green I dye
A)Biotin
B)SYBR Green II dye
C)Propidium iodide
D)SYBR Green I dye

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
9
Reverse transcriptase was discovered in
A)retroviruses.
B)adenoviruses.
C)herpes viruses.
D)Coxsackie viruses.
E)polioviruses.
A)retroviruses.
B)adenoviruses.
C)herpes viruses.
D)Coxsackie viruses.
E)polioviruses.

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
10
Which is more sensitive, nucleic acid amplification techniques or nucleic acid probe techniques?
A)Nucleic acid probe techniques.
B)Nucleic acid amplification techniques.
C)The sensitivities of the two techniques are approximately equal.
D)It depends on the particular technique as some amplification techniques are more sensitive than others.
A)Nucleic acid probe techniques.
B)Nucleic acid amplification techniques.
C)The sensitivities of the two techniques are approximately equal.
D)It depends on the particular technique as some amplification techniques are more sensitive than others.

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
11
What advantage does DNA polymerase from Thermus aquaticus (Taq polymerase)have over DNA polymerase from E. coli?
A)It synthesizes DNA more accurately with fewer random nucleotide mismatches.
B)DNA gyrase does not have to be added to the mixture to unwind the DNA prior to synthesis.
C)It is more heat stable.
D)It synthesizes DNA more rapidly.
A)It synthesizes DNA more accurately with fewer random nucleotide mismatches.
B)DNA gyrase does not have to be added to the mixture to unwind the DNA prior to synthesis.
C)It is more heat stable.
D)It synthesizes DNA more rapidly.

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
12
Review the PCR protocol herein and identify its name. A single- stranded probe with internal complementary sequences folds on itself into a "hairpin" shape bringing a fluorescent molecule at one end of the strand in proximity to the quencher molecule at the other end of the strand. After PCR product is formed, during the next denaturation step the probe becomes single- stranded, binds to PCR target DNA, and separates the fluorescent molecule from the quencher molecule. PCR product is detected as increased fluorescence.
A)Molecular beacon PCR
B)5' RACE PCR
C)Reverse transcriptase PCR
D)Branched chain PCR
E)TaqMan PCR
A)Molecular beacon PCR
B)5' RACE PCR
C)Reverse transcriptase PCR
D)Branched chain PCR
E)TaqMan PCR

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
13
When making mRNA preparations for reverse transcriptase PCR, it is most important to eliminate contamination from
A)transfer RNA.
B)ribosomal RNA.
C)nucleolar RNA.
D)DNA.
E)All of the above.
A)transfer RNA.
B)ribosomal RNA.
C)nucleolar RNA.
D)DNA.
E)All of the above.

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
14
The three basic steps of polymerase chain reaction (PCR)are denaturation, primer annealing, and primer extension. Which specification listed below is correct?
A)Primer design-oligonucleotide of 15- 30 base pairs
B)Primer extension-10 minutes during each cycle
C)Primer annealing-95° C for 1 minute
D)Denaturation-50° C for 5 minutes
E)Primer extension-37° C when Taq polymerase is used
A)Primer design-oligonucleotide of 15- 30 base pairs
B)Primer extension-10 minutes during each cycle
C)Primer annealing-95° C for 1 minute
D)Denaturation-50° C for 5 minutes
E)Primer extension-37° C when Taq polymerase is used

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
15
Which of the following is true regarding agarose gel electrophoresis of DNA?
A)The loading dye usually contains a marker dye and a heavy molecule such as glycerol.
B)In general, good separation of DNA fragments in the 1000-20,000 base pair size range can be achieved.
C)The usual buffer used is potassium phosphate at a pH of 7.2.
D)The usual stain to visualize the DNA is propidium iodide.
A)The loading dye usually contains a marker dye and a heavy molecule such as glycerol.
B)In general, good separation of DNA fragments in the 1000-20,000 base pair size range can be achieved.
C)The usual buffer used is potassium phosphate at a pH of 7.2.
D)The usual stain to visualize the DNA is propidium iodide.

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
16
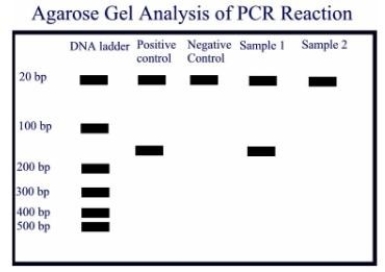
A PCR amplification was done on a positive control containing the template DNA, a negative control without the template DNA and two samples. Oligonucleotide primers of 21 base pairs were used, and the expected amplicon product is about 160 base pairs. Observe the agarose gel electrophoresis done on the amplicons, and give the most appropriate interpretations.
A)The assay must be repeated because the positive control has more than one amplicon.
B)The assay must be repeated because the amplicon is not the right size.
C)The assay must be repeated because there is evidence of "primer dimer" formation.
D)Sample 1 is positive for the template DNA, and sample 2 is negative for the template DNA.
E)The assay must be repeated because the negative control is not negative.

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
17
The three basic steps of polymerase chain reaction (PCR)are denaturation, primer annealing, and primer extension. Which step requires DNA polymerase?
A)Primer extension
B)Denaturation
C)Primer annealing
D)All of the above
A)Primer extension
B)Denaturation
C)Primer annealing
D)All of the above

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
18
Although must be present in excess, if the concentration is too high, an increase in errors and/or nonspecific products will occur.
A)All four deoxynucleotides (dNTPs)
B)Magnesium ions
C)Primers
D)All of the above
A)All four deoxynucleotides (dNTPs)
B)Magnesium ions
C)Primers
D)All of the above

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
19
What is an advantage of nucleic acid sequence based amplification (NASBA)?
A)It is faster than PCR.
B)Primers are not necessary.
C)The reaction is isothermal.
D)Product can be detected in real time.
A)It is faster than PCR.
B)Primers are not necessary.
C)The reaction is isothermal.
D)Product can be detected in real time.

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
20
How does one assure that all PCR amplicons are double- stranded?
A)By lengthening the final primer extension step from 1-2 minutes to up to 10 minutes.
B)By adding DNA ligase to the mixture.
C)By holding the reaction mixture at annealing temperature for about 5 minutes at the end of the cycle.
D)By making sure the dNTPs in the mixture are in excess.
A)By lengthening the final primer extension step from 1-2 minutes to up to 10 minutes.
B)By adding DNA ligase to the mixture.
C)By holding the reaction mixture at annealing temperature for about 5 minutes at the end of the cycle.
D)By making sure the dNTPs in the mixture are in excess.

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
21
mRNA preparations that have been treated with diethylpyrocarbonate (DEPC)can usually stored several weeks in the freezer.

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
22
Reverse transcriptase PCR can only be done on pure preparations of messenger RNA as ribosomal RNA and transfer RNA interfere significantly.

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
23
What technique listed is best to use to detect a mutated tumor gene in a section of tumor tissue?
A)Branched chain PCR
B)Ligase chain reaction
C)Nucleic acid sequence based amplification
D)In situ amplification
A)Branched chain PCR
B)Ligase chain reaction
C)Nucleic acid sequence based amplification
D)In situ amplification

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
24
Real time PCR is not as fast as traditional PCR, but it is more sensitive.

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
25
Which of the following typically produces a false positive in in situ nucleic acid amplification?
A)Incorporation of labeled nucleotides into DNA when DNA polymerase is repairing damaged DNA
B)Poor thermal conductance into cells
C)Low copy number of target sequence
D)Poor cell permeability
A)Incorporation of labeled nucleotides into DNA when DNA polymerase is repairing damaged DNA
B)Poor thermal conductance into cells
C)Low copy number of target sequence
D)Poor cell permeability

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
26
A standard PCR assay uses (how many?)primer(s).

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
27
Which amplification technique listed is best to use if one wishes to characterize the 5' end of an mRNA transcript?
A)3' RACE
B)Nucleic acid sequence based amplification (NASBA)
C)Branched chain DNA detection
D)Ligase chain reaction
E)5' RACE
A)3' RACE
B)Nucleic acid sequence based amplification (NASBA)
C)Branched chain DNA detection
D)Ligase chain reaction
E)5' RACE

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
28
Most nucleic acid amplification techniques amplify and detect target DNA, but in branched chain detection a labeled probe attaches to the target DNA and the label is amplified for detection.

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
29
The product of a PCR reaction is called an oligonucleotide.

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
30
Which amplification technique listed is best to use if one does not possess a thermal cycler?
A)Branched chain DNA detection
B)Ligase chain reaction
C)In situ amplification
D)5' RACE
E)Nucleic acid sequence based amplification (NASBA)
A)Branched chain DNA detection
B)Ligase chain reaction
C)In situ amplification
D)5' RACE
E)Nucleic acid sequence based amplification (NASBA)

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
31
The instrument used to control the heating cycles in PCR is called a .

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
32
Using SYBR green as a detection dye in real time PCR is more specific than traditional PCR, but it is not as sensitive.

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
33
It is important for laboratory workers to wash their hands thoroughly prior to working with RNA extracts.

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
34
Since multiple genes or gene products may be important in a particular disease, branched chain nucleic acid detection was developed to detect more than one target.

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
35
RNA- dependent DNA polymerase is the proper name for the enzyme .

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
36
Exposing work areas and equipment to is a useful decontamination method as it will cross- link and/or nick nucleic acids, and they will not be able to be amplified in PCR reactions.

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
37
Since Taq polymerase cannot function without free magnesium ions (Mg++)and many chemical species can bind Mg++, a gross excess of Mg++ must always be added to any PCR reaction mixture.

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
38
What technique combines PCR with in situ hybridization?
A)Branched chain PCR
B)In situ amplification
C)3' RACE
D)Nucleic acid sequence based amplification
E)5' RACE
A)Branched chain PCR
B)In situ amplification
C)3' RACE
D)Nucleic acid sequence based amplification
E)5' RACE

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
39
A standard reverse transcriptase PCR assay uses _ _ (how many?)primer(s)to convert mRNA to cDNA.

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
40
Even a small amount of contaminating nucleic acid can cause a false positive PCR result.

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
41
Explain how reverse transcriptase PCR can be used to accurately quantify RNA in different samples.

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
42
Describe the principle of TaqMan real- time PCR.

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
43
List ways in which contamination of PCR reactions can be minimized.

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
44
Discuss the factors that must be considered when designing primers for a PCR reaction.

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck
45
How can one quantify the amount of nucleic acid that is present in a sample and determine its purity?

Unlock Deck
Unlock for access to all 45 flashcards in this deck.
Unlock Deck
k this deck


