Deck 7: Dna Structure and Replication
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/57
Play
Full screen (f)
Deck 7: Dna Structure and Replication
1
If complementary DNA strands were arranged in a parallel manner, what would you expect to see?
A) The phosphodiester backbones would be too close and repel one another.
B) There would be no discernable difference between DNA strands aligned in a parallel versus an antiparallel manner.
C) Complementary nucleotides would line up normally; but fewer hydrogen bonds would form, so the strands could be more easily pulled apart.
D) Some regions of the two strands may form atypical hydrogen bonds, but the overall structure of the two DNA strands would not be stable.
E) Complementary nucleotides would be attracted to each other, forming ionic bonds that would make the helix stable but not uniform in width.
A) The phosphodiester backbones would be too close and repel one another.
B) There would be no discernable difference between DNA strands aligned in a parallel versus an antiparallel manner.
C) Complementary nucleotides would line up normally; but fewer hydrogen bonds would form, so the strands could be more easily pulled apart.
D) Some regions of the two strands may form atypical hydrogen bonds, but the overall structure of the two DNA strands would not be stable.
E) Complementary nucleotides would be attracted to each other, forming ionic bonds that would make the helix stable but not uniform in width.
D
2
What type of bond is formed between the hydroxyl group of one nucleotide and the phosphate group of an adjacent nucleotide, forming the sugar- phosphate backbone of DNA?
A) phosphodiester bond
B) glycosidic bond
C) hydrogen bond
D) ester linkage
E) ionic bond
A) phosphodiester bond
B) glycosidic bond
C) hydrogen bond
D) ester linkage
E) ionic bond
A
3
What is the difference between a nucleotide and a nucleoside?
A) Nucleotides are involved in eukaryotic DNA replication, while nucleosides are used in bacterial DNA replication.
B) A nucleoside with a phosphate ester linked to the sugar is a nucleotide.
C) Nucleosides contain only deoxyribose sugars.
D) Nucleosides are purines, while nucleotides are pyrimidines.
E) Nucleotides are found in DNA, while nucleosides are found in RNA.
A) Nucleotides are involved in eukaryotic DNA replication, while nucleosides are used in bacterial DNA replication.
B) A nucleoside with a phosphate ester linked to the sugar is a nucleotide.
C) Nucleosides contain only deoxyribose sugars.
D) Nucleosides are purines, while nucleotides are pyrimidines.
E) Nucleotides are found in DNA, while nucleosides are found in RNA.
B
4
The following represents a DNA strand in the process of replication. The bottom sequence is that of DNA strand with polarity indicated and the top sequence represents the RNA primer. GGGGCCUUU
5' AAATCCCCGGAAACTAAAC 3'
Which of the following will be the first DNA nucleotide added to the primer?
A) T
B) U
C) C
D) G
E) A
5' AAATCCCCGGAAACTAAAC 3'
Which of the following will be the first DNA nucleotide added to the primer?
A) T
B) U
C) C
D) G
E) A

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
5
What is the DNA replication fork?
A) It is the DNA strand that wraps around DNA polymerase and serves as the template for the lagging strand.
B) It is the site where the DNA helix opens to two single DNA strands.
C) It is the site where the leading and lagging strands meet on chromosome.
D) It is the DNA bound inside the DNA polymerase active site.
E) It is the binding site on the chromosome for DNA polymerase.
A) It is the DNA strand that wraps around DNA polymerase and serves as the template for the lagging strand.
B) It is the site where the DNA helix opens to two single DNA strands.
C) It is the site where the leading and lagging strands meet on chromosome.
D) It is the DNA bound inside the DNA polymerase active site.
E) It is the binding site on the chromosome for DNA polymerase.

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
6
Why would DNA synthesis occur in both directions from an origin of replication?
A) DNA is a double helix and DNA synthesis is 5' to 3'.
B) Replication- associated proteins bind at two replication forks.
C) It allows DNA strands to overlap when reaching another origin of replication.
D) Leading strands extend from one side of the origin while lagging strand extends from the other side of the origin.
E) DNA direction synthesis is random and sometimes extends in one direction from an origin of replication and sometimes in the other direction.
A) DNA is a double helix and DNA synthesis is 5' to 3'.
B) Replication- associated proteins bind at two replication forks.
C) It allows DNA strands to overlap when reaching another origin of replication.
D) Leading strands extend from one side of the origin while lagging strand extends from the other side of the origin.
E) DNA direction synthesis is random and sometimes extends in one direction from an origin of replication and sometimes in the other direction.

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
7
What is the difference between a ribonucleic acid and a deoxyribonucleic acid?
A) Deoxyribonucleic acids have a 2'H instead of the 2'OH found in ribonucleic acids.
B) Deoxyribonucleic acids are used to build nucleic acid strands, while ribonucleic acids are used exclusively in enzymatic reactions.
C) Ribonucleic acids have a 3'OH, while deoxyribonucleic acids have a 3'H.
D) Deoxyribonucleic acids have fewer oxygen groups and so are found only in monophosphate form in the cell, while ribonucleic acids have more oxygen groups and are found only in triphosphate forms.
E) Ribonucleic acid include A, T, G, and C, while deoxyribonucleic acids include A, U, G, and C.
A) Deoxyribonucleic acids have a 2'H instead of the 2'OH found in ribonucleic acids.
B) Deoxyribonucleic acids are used to build nucleic acid strands, while ribonucleic acids are used exclusively in enzymatic reactions.
C) Ribonucleic acids have a 3'OH, while deoxyribonucleic acids have a 3'H.
D) Deoxyribonucleic acids have fewer oxygen groups and so are found only in monophosphate form in the cell, while ribonucleic acids have more oxygen groups and are found only in triphosphate forms.
E) Ribonucleic acid include A, T, G, and C, while deoxyribonucleic acids include A, U, G, and C.

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
8
The process by which Pneumococcus transfers DNA between living type RII and heat- killed type SIII cells is known as _.
A) transformation
B) ligation
C) conjugation
D) transduction
E) replication
A) transformation
B) ligation
C) conjugation
D) transduction
E) replication

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
9
What types of bonds are formed between complementary DNA bases?
A) phosphodiester bonds
B) ionic bonds
C) hydrogen bonds
D) covalent bonds
E) glycosidic bonds
A) phosphodiester bonds
B) ionic bonds
C) hydrogen bonds
D) covalent bonds
E) glycosidic bonds

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
10
What is one difference between DNA replication in bacteria versus eukaryotes?
A) Bacterial chromosomes are replicated bi- directionally, while eukaryotic chromosomes are replicated in one direction.
B) The process is identical in bacterial and eukaryotic DNA replication.
C) Eukaryotic chromosomes have many origins of replication, while bacteria have only one origin of replication.
D) Eukaryotic chromosomes are replicated bi- directionally, while bacterial chromosomes are replicated in one direction.
E) Eukaryotic chromosomes have many origins of replication and replicate bi- directionally, while bacteria have only one origin of replication and replicate uni- directionally.
A) Bacterial chromosomes are replicated bi- directionally, while eukaryotic chromosomes are replicated in one direction.
B) The process is identical in bacterial and eukaryotic DNA replication.
C) Eukaryotic chromosomes have many origins of replication, while bacteria have only one origin of replication.
D) Eukaryotic chromosomes are replicated bi- directionally, while bacterial chromosomes are replicated in one direction.
E) Eukaryotic chromosomes have many origins of replication and replicate bi- directionally, while bacteria have only one origin of replication and replicate uni- directionally.

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
11
In a single strand of DNA, what fraction of the nucleotides in a molecule are cytosine and thymine (% C and % T added together)?
A) 25%
B) 75%
C) 50%
D) 100%
E) It depends on the DNA sequence
A) 25%
B) 75%
C) 50%
D) 100%
E) It depends on the DNA sequence

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
12
Based on the following replication bubble, which of these statements is true? 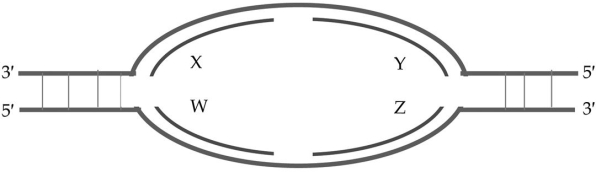
A) W and Z are leading strands, X and Y are lagging strands
B) X and Z are leading strands, W and Y are lagging strands
C) X and Y are leading strands, W and Z are lagging strands
D) W and Y are leading strands, X and Z are lagging strands
E) X and W are leading strands, Y and Z are lagging strands

A) W and Z are leading strands, X and Y are lagging strands
B) X and Z are leading strands, W and Y are lagging strands
C) X and Y are leading strands, W and Z are lagging strands
D) W and Y are leading strands, X and Z are lagging strands
E) X and W are leading strands, Y and Z are lagging strands

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
13
In the Hershey- Chase experiment, bacteriophages were produced in either 32P- containing or 35S- containing medium. Where were these isotopes eventually detected when the radioactively- labeled bacteriophages were introduced to a fresh bacterial culture?
A) The 32P was associated with the culture medium and 35S was associated with the phage particles.
B) Both 32P and 35S were associated with the bacterial cells.
C) The 32P was associated with the culture medium and 35S was associated with the bacterial cells.
D) Both 32P and 35S were associated with the phage particles.
E) The 32P was associated with the bacterial cells and 35S was associated with the phage particles.
A) The 32P was associated with the culture medium and 35S was associated with the phage particles.
B) Both 32P and 35S were associated with the bacterial cells.
C) The 32P was associated with the culture medium and 35S was associated with the bacterial cells.
D) Both 32P and 35S were associated with the phage particles.
E) The 32P was associated with the bacterial cells and 35S was associated with the phage particles.

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
14
If there is 24% guanine in a DNA molecule, then there is cytosine.
A) 48%
B) 24%
C) 26%
D) 52%
E) Not possible to determine
A) 48%
B) 24%
C) 26%
D) 52%
E) Not possible to determine

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
15
A portion of one strand of DNA has the sequence 5' AATGGCTTA 3'. If this strand is used as a template for DNA replication, which of the following correctly depicts the sequence of the newly synthesized strand in the direction in which it will be synthesized?
A) 5' TAAGCCATT 3'
B) 5' AATGGCTTA 3'
C) 3' AATGGCTTA 5'
D) 5' TTACCGAAT 3'
E) 3' TTACCGAAT 5'
A) 5' TAAGCCATT 3'
B) 5' AATGGCTTA 3'
C) 3' AATGGCTTA 5'
D) 5' TTACCGAAT 3'
E) 3' TTACCGAAT 5'

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
16
Why do origins of replication in various bacteria have conserved DNA consensus sequences?
A) Fewer mutations in the DNA occur at the very beginning of replication at the origin, hence why they are conserved as consensus sequences.
B) It is more efficient to use the same sequence multiple times in different genomes than to change it.
C) The consensus sequence allows the origin to wrap into a circle and recruit replication initiation proteins.
D) Origins need to be recognized by replication initiation proteins to open up their AT- rich regions.
E) Consensus sequences are found throughout bacterial genomes and so would be found at the origin as well.
A) Fewer mutations in the DNA occur at the very beginning of replication at the origin, hence why they are conserved as consensus sequences.
B) It is more efficient to use the same sequence multiple times in different genomes than to change it.
C) The consensus sequence allows the origin to wrap into a circle and recruit replication initiation proteins.
D) Origins need to be recognized by replication initiation proteins to open up their AT- rich regions.
E) Consensus sequences are found throughout bacterial genomes and so would be found at the origin as well.

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
17
Which arrow(s) point(s) to the location of helicase in the diagram shown? 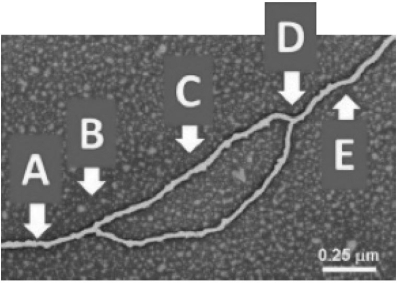
A) A
B) B
C) C
D) A and E
E) B and D

A) A
B) B
C) C
D) A and E
E) B and D

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
18
In their famous experiment, which of the following would Meselson and Stahl have observed after one cycle of replication in 14N medium if DNA replication were CONSERVATIVE?
A) An equal number of DNA molecules containing two 15N- DNA strands and DNA molecules containing two 14N- DNA strands
B) DNA molecules containing one strand of 15N- DNA and one strand of 14N- DNA
C) A mix of DNA molecules corresponding to A and B
D) DNA molecules containing two 15N- DNA strands only
E) DNA molecules containing two 14N- DNA strands only
A) An equal number of DNA molecules containing two 15N- DNA strands and DNA molecules containing two 14N- DNA strands
B) DNA molecules containing one strand of 15N- DNA and one strand of 14N- DNA
C) A mix of DNA molecules corresponding to A and B
D) DNA molecules containing two 15N- DNA strands only
E) DNA molecules containing two 14N- DNA strands only

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
19
Avery, Macleod, and McCarty expanded on Griffith's experiment to prove that DNA is the hereditary molecule required for transformation. What treatment of the heat- killed SIII bacteria extract resulted in the mouse LIVING?
A) Destruction of type SIII DNA with DNase
B) Destruction of type SIII lipids and polysaccharides
C) Destruction of type SIII RNA with RNase
D) Destruction of type SIII proteins with protease
E) Use of the control group (null treatment), in which all components are intact
A) Destruction of type SIII DNA with DNase
B) Destruction of type SIII lipids and polysaccharides
C) Destruction of type SIII RNA with RNase
D) Destruction of type SIII proteins with protease
E) Use of the control group (null treatment), in which all components are intact

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
20
Which of the following are classified as pyrimidines?
A) thymine and cytosine
B) adenine and uracil
C) guanine and cytosine
D) adenine and guanine
E) adenine and thymine
A) thymine and cytosine
B) adenine and uracil
C) guanine and cytosine
D) adenine and guanine
E) adenine and thymine

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
21
If Single- Stranded Binding protein (SSB) is NOT present during DNA replication, what would you expect to see?
A) The replisome complex would not assemble on the oriC region.
B) SSB carries the helicase protein to the open region of DNA, so hydrolysis and strand separation will not occur.
C) Helicase activity is inhibited, so DNA strands cannot be separated.
D) SSB prevents reannealing of the separated strands, so strands would quickly reanneal and DNA replication cannot proceed.
E) The DNA cannot bend, so hydrogen bonds in the 13- mer region of oriC remain intact.
A) The replisome complex would not assemble on the oriC region.
B) SSB carries the helicase protein to the open region of DNA, so hydrolysis and strand separation will not occur.
C) Helicase activity is inhibited, so DNA strands cannot be separated.
D) SSB prevents reannealing of the separated strands, so strands would quickly reanneal and DNA replication cannot proceed.
E) The DNA cannot bend, so hydrogen bonds in the 13- mer region of oriC remain intact.

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
22
Why are telomeres problematic for eukaryotic chromosome replication?
A) Telomerase is more error- prone than the normal DNA Polymerase.
B) Removal of the lagging strand primer leaves a gap in the one of the strand's DNA sequences.
C) The T loop blocks formation of primers on the lagging strand.
D) They are highly repetitive and thus hard to replicate correctly.
E) Maintaining very long telomeres promotes cancer cell formation.
A) Telomerase is more error- prone than the normal DNA Polymerase.
B) Removal of the lagging strand primer leaves a gap in the one of the strand's DNA sequences.
C) The T loop blocks formation of primers on the lagging strand.
D) They are highly repetitive and thus hard to replicate correctly.
E) Maintaining very long telomeres promotes cancer cell formation.

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
23
Which of the following would you find in a Sanger sequencing reaction but not in a polymerase chain reaction?
A) dNTPs
B) DNA primer
C) DNA polymerase
D) DNA template
E) ddNTPs
A) dNTPs
B) DNA primer
C) DNA polymerase
D) DNA template
E) ddNTPs

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
24
Where would you expect to find telomerase activity?
A) At the centromere of a chromosome in a healthy eukaryotic reproductive cell.
B) At the end of a chromosome in a normal healthy eukaryotic body (somatic) cell.
C) On the leading strand of DNA in a normal bacterial cell.
D) On the lagging strand of DNA in a normal bacterial cell.
E) At the end of a chromosome in a cancerous eukaryotic body cell.
A) At the centromere of a chromosome in a healthy eukaryotic reproductive cell.
B) At the end of a chromosome in a normal healthy eukaryotic body (somatic) cell.
C) On the leading strand of DNA in a normal bacterial cell.
D) On the lagging strand of DNA in a normal bacterial cell.
E) At the end of a chromosome in a cancerous eukaryotic body cell.

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
25
Why are leading and lagging strand primers removed rather than joined with Okazaki fragments?
A) They lack the 3' OH that would permit them to be covalently linked to the Okazaki fragments.
B) The primers do not hydrogen bond to the template strand correctly.
C) They contain nucleotides with 2'OH groups, and are targeted for excision by DNA Polymerase.
D) They have to be removed for leading and lagging strand synthesis to begin.
E) Primers are too short to efficiently bind to Okazaki fragments.
A) They lack the 3' OH that would permit them to be covalently linked to the Okazaki fragments.
B) The primers do not hydrogen bond to the template strand correctly.
C) They contain nucleotides with 2'OH groups, and are targeted for excision by DNA Polymerase.
D) They have to be removed for leading and lagging strand synthesis to begin.
E) Primers are too short to efficiently bind to Okazaki fragments.

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
26
Which of the following temperature cycles would you expect to see in a standard polymerase chain reaction (from denaturation to annealing to extension)?
A) 95° -55° -72°
B) 45° -72° -95°
C) 95° -72° -95°
D) 75° -95° -45°
E) 95° -72° -55°
A) 95° -55° -72°
B) 45° -72° -95°
C) 95° -72° -95°
D) 75° -95° -45°
E) 95° -72° -55°

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
27
You identify a cell in which DNA polymerase III is functional, but it seems to exhibit extremely low processivity. This is likely a defect in what structure?
A) the topoisomerase enzyme
B) the pol III holoenzyme
C) the sliding clamp
D) the clamp loader
E) the t proteins
A) the topoisomerase enzyme
B) the pol III holoenzyme
C) the sliding clamp
D) the clamp loader
E) the t proteins

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
28
What is removed from dNTPs to provide the energy required for DNA polymerase to catalyze DNA strand elongation?

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
29
DNA helicase inhibitors are well studied as potential drug targets. What would you expect to see if DNA helicase activity is inhibited?
A) Helicase prevents reannealing of the separated strands, so strands would quickly reanneal and DNA replication cannot proceed.
B) The DNA cannot bend, so hydrogen bonds in the 13- mer region of oriC remain intact.
C) Helicase carries the SSB protein to the open region of DNA, so hydrolysis and strand separation will not occur.
D) Helicase catalyzes ATP hydrolysis and DNA strands separation, so the helix cannot be unwound and strands will not separate.
E) The replisome complex would not assemble on the oriC region.
A) Helicase prevents reannealing of the separated strands, so strands would quickly reanneal and DNA replication cannot proceed.
B) The DNA cannot bend, so hydrogen bonds in the 13- mer region of oriC remain intact.
C) Helicase carries the SSB protein to the open region of DNA, so hydrolysis and strand separation will not occur.
D) Helicase catalyzes ATP hydrolysis and DNA strands separation, so the helix cannot be unwound and strands will not separate.
E) The replisome complex would not assemble on the oriC region.

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
30
What is the purpose of the 3'- to- 5' exonuclease activity of DNA Polymerase?
A) Removing mismatched nucleotides at the polymerase active site increases the rate of synthesis in the 5' to 3' direction.
B) Degrade excessive nucleotides on Okazaki fragments.
C) Remove mismatched nucleotides in the template strand.
D) Some polymerases do not have exonuclease activity.
E) Remove mismatched nucleotides in the newly synthesized strand.
A) Removing mismatched nucleotides at the polymerase active site increases the rate of synthesis in the 5' to 3' direction.
B) Degrade excessive nucleotides on Okazaki fragments.
C) Remove mismatched nucleotides in the template strand.
D) Some polymerases do not have exonuclease activity.
E) Remove mismatched nucleotides in the newly synthesized strand.

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
31
The extraordinary accuracy of the DNA polymerase III enzyme lies in its ability to "proofread" newly synthesized DNA, a function of the enzyme's .
A) 3'- to- 5' helicase activity
B) 5'- to- 3' polymerase activity
C) 3'- to- 5' exonuclease activity
D) 5'- to- 3' exonuclease activity
E) 3'- to- 5' polymerase activity
A) 3'- to- 5' helicase activity
B) 5'- to- 3' polymerase activity
C) 3'- to- 5' exonuclease activity
D) 5'- to- 3' exonuclease activity
E) 3'- to- 5' polymerase activity

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
32
Which functional groups have been altered in a ddNTP compared to a dNTP?
A) The ddNTPs have a 2' H and a 3' OH, while dNTPs have a 2' H and a 3' H.
B) The ddNTPs have a 2' OH and a 3' H, while dNTPs have a 2' H and a 3' OH.
C) The ddNTPs have a 2' H and a 3' H, while dNTPs have a 2' H and a 3' OH.
D) The ddNTPs have a 2' H and a 3' H, while dNTPs have a 2' OH and a 3' OH.
E) The ddNTPs have a 2' OH and a 3' OH, while dNTPs have a 2' H and a 3' H.
A) The ddNTPs have a 2' H and a 3' OH, while dNTPs have a 2' H and a 3' H.
B) The ddNTPs have a 2' OH and a 3' H, while dNTPs have a 2' H and a 3' OH.
C) The ddNTPs have a 2' H and a 3' H, while dNTPs have a 2' H and a 3' OH.
D) The ddNTPs have a 2' H and a 3' H, while dNTPs have a 2' OH and a 3' OH.
E) The ddNTPs have a 2' OH and a 3' OH, while dNTPs have a 2' H and a 3' H.

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
33
What are the regions where DNA- binding proteins can make direct contact with exposed nucleotides?

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
34
Why are next generation DNA sequencing technologies known as sequencing- by- synthesis?
A) Numerous synthesized fragments of DNA are sequenced to determine which nucleotides were incorporated.
B) Incorporated nucleotides are determined while they are being added to a growing DNA strand.
C) RNA molecules are synthesized off of the DNA templates and the incorporated nucleotides are then determined.
D) The complete DNA strands are synthesized then sequenced.
E) The sequencing occurs during S phase of the cell cycle.
A) Numerous synthesized fragments of DNA are sequenced to determine which nucleotides were incorporated.
B) Incorporated nucleotides are determined while they are being added to a growing DNA strand.
C) RNA molecules are synthesized off of the DNA templates and the incorporated nucleotides are then determined.
D) The complete DNA strands are synthesized then sequenced.
E) The sequencing occurs during S phase of the cell cycle.

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
35
What is the overall direction of DNA strand elongation?

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
36
What proteins bind the DNA sequences found at the origins of replication?

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
37
What are the three parts of a DNA nucleotide?

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
38
During DNA replication in E. coli, how does the cell prevent the DNA strands from following their natural tendency to seek maximum stability and reanneal?

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
39
Based on Griffith's results, what would you expect if you injected both heat- killed type RII and living type SIII?

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
40
What is required for DNA polymerase to initiate DNA strand synthesis?
A) a short DNA primer synthesized by the enzyme primase
B) a short RNA primer synthesized by the enzyme primase
C) ATP and a short RNA primer synthesized by the enzyme topoisomerase
D) ATP and a short DNA primer synthesized by the enzyme topoisomerase
E) DNA polymerase initiates DNA strand synthesis without requiring any additional enzymes.
A) a short DNA primer synthesized by the enzyme primase
B) a short RNA primer synthesized by the enzyme primase
C) ATP and a short RNA primer synthesized by the enzyme topoisomerase
D) ATP and a short DNA primer synthesized by the enzyme topoisomerase
E) DNA polymerase initiates DNA strand synthesis without requiring any additional enzymes.

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
41
Describe the DNA helix structure proposed by Watson and Crick. What is the diameter of the helix and the distance between bases? What types of bonds are involved in stabilizing the DNA backbone, and what types of bonds are found between complementary base pairs? Why would a sequence containing high concentrations of G and C require more energy to break the bonds?

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
42
What structure in E. coli delivers primase and accessory proteins to the oriC site to synthesize the primer needed for DNA replication?

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
43
What characteristic of the polymerase isolated from Thermus aquaticus makes it unique and highly useful for maintaining the enzyme efficiency during PCR?

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
44
Which enzyme, a combination of several proteins and a molecule of RNA, acts as the template for the telomere repeat DNA sequence?

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
45
What are the three steps of each PCR cycle?

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
46
What did Okazaki's discovery of the short segments of DNA reveal about DNA replication on the lagging strand?

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
47
What is the term for the daughter strand synthesized continuously during DNA replication?

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
48
The major and minor grooves of DNA are features of the helix that can be attributed to
, which involves tight packing of DNA bases in a duplex.
, which involves tight packing of DNA bases in a duplex.

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
49
Many antibacterial agents and chemotherapy agents act as topoisomerase inhibitors. Why would inhibition of topoisomerase cause cell death in bacterial and eukaryotic cells?

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
50
The large, complex aggregation of proteins and enzymes that assembles at the replication fork is known as the .

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
51
DNA polymerase requires a primer sequence to provide a functional group to which the new DNA nucleotide can be added.

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
52
is the term for a multiprotein complex in which a core enzyme is associated with additional protein components leading to its function.

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
53
How might you choose a region of DNA for a PCR primer so as to increase the temperature necessary for primer annealing (to minimize nonspecific PCR products)?

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
54
Briefly describe the Meselson and Stahl experiment that indicated that DNA replication is semiconservative. How would their results have differed if DNA replication were actually conservative in nature?

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
55
must be added to a PCR to direct the polymerase where to begin synthesis.

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
56
Describe the function of the oriC. What do you predict will happen to the cell if 100 bp between the 13- mer and 9- mer sequence is deleted as indicated in the figure?
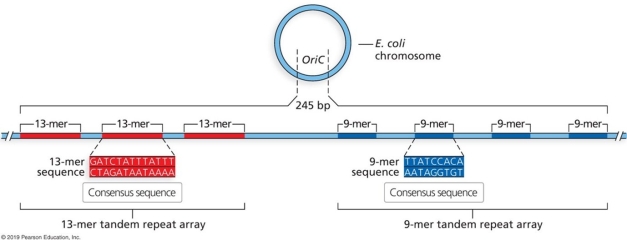


Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck
57
Comparison of conserved sequences among related species usually leads to the identification of , which illustrate the nucleotides most often found at each position of DNA in the conserved region.

Unlock Deck
Unlock for access to all 57 flashcards in this deck.
Unlock Deck
k this deck


