Deck 20: Dna Replication, Repair, and Recombination
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/76
Play
Full screen (f)
Deck 20: Dna Replication, Repair, and Recombination
1
Enzymes that remain bound to their nascent chains through many polymerization steps are said to be .
A) constitutive
B) distributive
C) conjugated
D) processive
A) constitutive
B) distributive
C) conjugated
D) processive
processive
2
E. coli DNA polymerase catalyzes polymerization in the directions).
A) 5ʹ → 3ʹ
B) 3ʹ → 5ʹ
C) 2ʹ → 5ʹ
D) both 5ʹ → 3ʹ and 3ʹ → 5ʹ
E) All of the above
A) 5ʹ → 3ʹ
B) 3ʹ → 5ʹ
C) 2ʹ → 5ʹ
D) both 5ʹ → 3ʹ and 3ʹ → 5ʹ
E) All of the above
5ʹ → 3ʹ
3
Which DNA polymerase is most responsible for chain elongation in E. coli?
A) DNA polymerase I
B) DNA polymerase II
C) DNA polymerase III
D) DNA polymerase IV
A) DNA polymerase I
B) DNA polymerase II
C) DNA polymerase III
D) DNA polymerase IV
DNA polymerase III
4
The protein machine that carries out DNA replication is called .
A) DNA polymerase
B) the replication fork
C) the Klenow fragment
D) the replisome
A) DNA polymerase
B) the replication fork
C) the Klenow fragment
D) the replisome

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
5
Nick translation is a process for
A) filling in and sealing the gap in DNA after removal of the RNA primer.
B) synthesis of the protein for primase from mRNA.
C) removal of the RNA primer by DNA polymerase I.
D) removal of Okazaki fragments.
A) filling in and sealing the gap in DNA after removal of the RNA primer.
B) synthesis of the protein for primase from mRNA.
C) removal of the RNA primer by DNA polymerase I.
D) removal of Okazaki fragments.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
6
Approximately base pairs are added per second during DNA in vivo replication in E. coli.
A) 10
B) 100
C) 1000
D) 1,000,000
A) 10
B) 100
C) 1000
D) 1,000,000

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
7
Polymerization of the lagging strand is catalyzed by .
A) a second core complex on the same polymerase holoenzyme that polymerizes the leading strand
B) a completely separate replisome from that which polymerizes the leading strand
C) DNA polymerase I holoenzyme
D) the same polymerase holoenzyme that polymerized the leading strand, but with opposite directionality
A) a second core complex on the same polymerase holoenzyme that polymerizes the leading strand
B) a completely separate replisome from that which polymerizes the leading strand
C) DNA polymerase I holoenzyme
D) the same polymerase holoenzyme that polymerized the leading strand, but with opposite directionality

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
8
It takes about the same amount of time for eukaryotic chromosomes to be replicated in vivo as it does for E. coli. Why is this true?
A) Eukaryotes have multiple origins of replication; E. coli has one unique origin.
B) Most chromosomes are approximately the same size.
C) Eukaryotic replication occurs at a faster rate, enabling replication of the larger chromosomes in about the same amount of time.
D) The replication of DNA in eukaryotes is faster due to a quicker, more accurate repair of mismatches.
A) Eukaryotes have multiple origins of replication; E. coli has one unique origin.
B) Most chromosomes are approximately the same size.
C) Eukaryotic replication occurs at a faster rate, enabling replication of the larger chromosomes in about the same amount of time.
D) The replication of DNA in eukaryotes is faster due to a quicker, more accurate repair of mismatches.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
9
The RNA primer at the beginning of each Okazaki fragment is removed by
A) DNA polymerase I.
B) DNA polymerase II.
C) DNA polymerase III.
D) 3ʹ → 5ʹ exonuclease subunit of DNA polymerase III.
A) DNA polymerase I.
B) DNA polymerase II.
C) DNA polymerase III.
D) 3ʹ → 5ʹ exonuclease subunit of DNA polymerase III.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
10
The 3ʹ → 5ʹ exonuclease activity of E. coli DNA polymerase III accounts for the of polymerization.
A) low error rate
B) high speed
C) directionality
D) All of the above
A) low error rate
B) high speed
C) directionality
D) All of the above

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
11
The incorporation of new nucleotides into a growing strand of DNA occurs during .
A) the elongation stage
B) nucleation
C) initiation
D) assembly of the protein machine
A) the elongation stage
B) nucleation
C) initiation
D) assembly of the protein machine

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
12
How many replisomes are required for one complete round of DNA replication in E. coli?
A) one
B) two
C) four
D) more than 4
A) one
B) two
C) four
D) more than 4

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
13
During the elongation stage
A) replication proteins assemble at the DNA replication site.
B) DNA is replicated semiconservatively.
C) the protein machine disassembles.
D) replication ends.
A) replication proteins assemble at the DNA replication site.
B) DNA is replicated semiconservatively.
C) the protein machine disassembles.
D) replication ends.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
14
DNA polymerase III requires a/an for synthesis of DNA to occur.
A) DNA template
B) RNA primer
C) free 3ʹ-OH end of a DNA strand
D) All of the above
A) DNA template
B) RNA primer
C) free 3ʹ-OH end of a DNA strand
D) All of the above

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
15
The e subunit of DNA polymerase III is responsible for its activity.
A) 5ʹ → 3ʹ polymerase
B) sliding clamp
C) 3ʹ → 5ʹ exonuclease proof reading)
D) ribosome assembly
A) 5ʹ → 3ʹ polymerase
B) sliding clamp
C) 3ʹ → 5ʹ exonuclease proof reading)
D) ribosome assembly

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
16
DNA that has been liberally labeled with radioactive 14C is used as the template for replication. Replication is carried out in a medium containing only unlabeled nucleotides. After two rounds of replication, what percent of DNA molecules are still labeled?
A) 25%
B) 50%
C) 75%
D) 100%
A) 25%
B) 50%
C) 75%
D) 100%

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
17
The replication of DNA is because .
A) conservative; one strand of parental DNA is retained in each daughter DNA
B) conservative; each daughter molecule has two new strands copied from the parental DNA template
C) semiconservative; one strand of parental DNA is retained in each daughter DNA
D) semiconservative; each daughter molecule has two new strands copied from the parental DNA template
A) conservative; one strand of parental DNA is retained in each daughter DNA
B) conservative; each daughter molecule has two new strands copied from the parental DNA template
C) semiconservative; one strand of parental DNA is retained in each daughter DNA
D) semiconservative; each daughter molecule has two new strands copied from the parental DNA template

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
18
Meselson and Stahlʹs experiments involved growing E. coli on a medium containing 15N heavy nitrogen) which increased the density of the bacterial DNA. Cells were then grown on a medium containing only 14N. DNA samples were obtained for analysis after one, two and three rounds of replication generations) and centrifuged on a density gradient. The double-helical DNA forms bands in the gradient depending on its density. How many bands did Meselson and Stahl see after two generation of cells were grown on the 14N medium?
A) One with DNA containing both 15N and 14N.
B) Two, one with DNA containing only 15N and one with DNA containing only 14N.
C) Two, one with DNA containing both 15N and 14N and one with DNA containing only 14N.
D) Three, one with DNA containing only 15N, one with DNA containing both 15N and 14N and one with DNA containing only 14N.
A) One with DNA containing both 15N and 14N.
B) Two, one with DNA containing only 15N and one with DNA containing only 14N.
C) Two, one with DNA containing both 15N and 14N and one with DNA containing only 14N.
D) Three, one with DNA containing only 15N, one with DNA containing both 15N and 14N and one with DNA containing only 14N.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
19
The processivity of E. coli DNA polymerase III holoenzyme accounts for all of the following except
A) the relatively small number of enzymes needed to replicate the entire chromosome.
B) the directionality of polymerization.
C) the rapid rate of polymerization.
D) the ability of a single holoenzyme to add many nucleotides to a growing DNA chain.
A) the relatively small number of enzymes needed to replicate the entire chromosome.
B) the directionality of polymerization.
C) the rapid rate of polymerization.
D) the ability of a single holoenzyme to add many nucleotides to a growing DNA chain.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
20
At the normal rate of replication, about how many minutes does it take for the entire E. coli chromosome 4.6 × 103 kbp) to be duplicated?
A) 5
B) 20
C) 40
D) 80
A) 5
B) 20
C) 40
D) 80

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
21
Functions of DNA polymerases and accessory proteins in eukaryotic DNA replication include
A) PCNA protein, a sliding clamp unit for DNA.
B) β- DNA polymerase, for repair of damaged DNA.
C) RPC protein, and equivalent of SSB proteins in bacteria.
D) RPC protein, used for mitochondrial DNA synthesis.
E) All of the above
A) PCNA protein, a sliding clamp unit for DNA.
B) β- DNA polymerase, for repair of damaged DNA.
C) RPC protein, and equivalent of SSB proteins in bacteria.
D) RPC protein, used for mitochondrial DNA synthesis.
E) All of the above

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
22
Which of the following is mismatched?
A) ter : Tus
B) dnaA : DnaA
C) Ori : primer
D) Tus : helicase inhibitor
A) ter : Tus
B) dnaA : DnaA
C) Ori : primer
D) Tus : helicase inhibitor

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
23
In photoreactivation, DNA damage caused by UV light is reversed by DNA photolyase which
A) is activated by UV light to form a complex.
B) removes nicks in the DNA backbone.
C) forms a complex with thymine dimers which absorbs visible light and reverses the dimerization.
D) binds to the adenine bases opposite the thymine dimer.
E) All of the above
A) is activated by UV light to form a complex.
B) removes nicks in the DNA backbone.
C) forms a complex with thymine dimers which absorbs visible light and reverses the dimerization.
D) binds to the adenine bases opposite the thymine dimer.
E) All of the above

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
24
Fragments on the lagging strand are joined by the enzyme .
A) DNA polymerase I
B) DNA synthase
C) DNA primosome
D) DNA ligase
A) DNA polymerase I
B) DNA synthase
C) DNA primosome
D) DNA ligase

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
25
Why does E. coli need both DNA polymerse III and DNA polymerase I?
A) DNA polymerase III lacks the 5ʹ → 3ʹ exonuclease activity needed to remove RNA primers.
B) Each polymerase is specific for only one strand of DNA. DNA polymerase III acts only on the leading strand, and DNA polymerase I acts only on the lagging strand.
C) DNA polymerase I is needed to join the Okazaki fragments.
D) The DNA replication is bidirectional; one polymerase is used for each direction.
E) Only DNA polymerase I has proofreading ability.
A) DNA polymerase III lacks the 5ʹ → 3ʹ exonuclease activity needed to remove RNA primers.
B) Each polymerase is specific for only one strand of DNA. DNA polymerase III acts only on the leading strand, and DNA polymerase I acts only on the lagging strand.
C) DNA polymerase I is needed to join the Okazaki fragments.
D) The DNA replication is bidirectional; one polymerase is used for each direction.
E) Only DNA polymerase I has proofreading ability.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
26
Okazaki fragments are .
A) the smallest subunits of DNA polymerase III
B) short stretches of DNA formed on the lagging strand
C) short RNA primers needed for initiation of polymerization
D) fragments of DNA polymerase I that lack 5ʹ → 3ʹ exonuclease activity
A) the smallest subunits of DNA polymerase III
B) short stretches of DNA formed on the lagging strand
C) short RNA primers needed for initiation of polymerization
D) fragments of DNA polymerase I that lack 5ʹ → 3ʹ exonuclease activity

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
27
The main difference between prokaryotic and eukaryotic DNA replication mechanisms is
A) no Okazaki fragments are made in eukaryotes.
B) both leading and lagging strands are continuous in eukaryotes.
C) no primer synthesis is needed in eukaryotes.
D) the replication fork moves slower in eukaryotes.
A) no Okazaki fragments are made in eukaryotes.
B) both leading and lagging strands are continuous in eukaryotes.
C) no primer synthesis is needed in eukaryotes.
D) the replication fork moves slower in eukaryotes.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
28
The best enzyme to use in the Sanger method for DNA sequence determination is
A) DNA polymerase I .
B) Klenow fragment of DNA polymerase I.
C) DNA polymerase from a high temperature bacterium.
D) SSB protein.
A) DNA polymerase I .
B) Klenow fragment of DNA polymerase I.
C) DNA polymerase from a high temperature bacterium.
D) SSB protein.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
29
Why is only one copy of DNA made per cell division cycle in eukaryotes despite the presence of multiple origins of replication?
A) There is only one origin of replication per chromosome.
B) The rate of elongation proceeds at a rate that allows only one copy to be made by the time the cell divides.
C) The activity of an S-phase protein kinase controls the formation of the pre-replication complex.
D) Primase is activated by phosphorylation only at the onset of S-phase by S-phase protein kinase.
A) There is only one origin of replication per chromosome.
B) The rate of elongation proceeds at a rate that allows only one copy to be made by the time the cell divides.
C) The activity of an S-phase protein kinase controls the formation of the pre-replication complex.
D) Primase is activated by phosphorylation only at the onset of S-phase by S-phase protein kinase.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
30
Which proteins are responsible for the unwinding of the double-stranded DNA during replication?
A) ligases
B) helicases
C) topoisomerases
D) primases
E) lyases
A) ligases
B) helicases
C) topoisomerases
D) primases
E) lyases

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
31
What is the function of the primosome?
A) Removal of RNA from the lagging strand.
B) Synthesis of short RNA fragments.
C) Assembly of DNA polymerase III holoenzyme.
D) Detection of the origin of replication.
A) Removal of RNA from the lagging strand.
B) Synthesis of short RNA fragments.
C) Assembly of DNA polymerase III holoenzyme.
D) Detection of the origin of replication.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
32
Which is not part of the replisome?
A) helicase DnaB
B) the primosome
C) gyrase
D) single-stranded binding protein
A) helicase DnaB
B) the primosome
C) gyrase
D) single-stranded binding protein

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
33
The reaction catalyzed by DNA photolyase is an example of what type of repair?
A) recombination
B) excision
C) mismatch
D) direct
A) recombination
B) excision
C) mismatch
D) direct

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
34
The sequence of genes and proteins used in the initiation and termination of DNA replication is
A) Tus → ter → origin → DnaA → dnaA
B) dnaA → DnaA → origin → Tus → ter
C) Origin → DnaA → dnaA → ter → Tus
D) DnaA → ter → Tus → origin → ter → dnaA
A) Tus → ter → origin → DnaA → dnaA
B) dnaA → DnaA → origin → Tus → ter
C) Origin → DnaA → dnaA → ter → Tus
D) DnaA → ter → Tus → origin → ter → dnaA

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
35
RNA primers are synthesized about once every .
A) millisecond
B) second
C) minute
D) 40 minutes
A) millisecond
B) second
C) minute
D) 40 minutes

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
36
What is a Klenow fragment?
A) A fragment of DNA polymerase I that lacks 5ʹ → 3ʹ exonuclease activity.
B) A topoisomerase responsible for unwinding DNA at the replication fork.
C) A portion of the replisome responsible for RNA primer synthesis.
D) Short stretches of DNA formed during polymerization of the lagging strand.
A) A fragment of DNA polymerase I that lacks 5ʹ → 3ʹ exonuclease activity.
B) A topoisomerase responsible for unwinding DNA at the replication fork.
C) A portion of the replisome responsible for RNA primer synthesis.
D) Short stretches of DNA formed during polymerization of the lagging strand.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
37
Which is not an activity for DNA polymerase I?
A) 5ʹ → 3ʹ polymerase activity
B) joining of Okazaki fragments
C) 5ʹ → 3ʹ exonuclease activity
D) nick translation
E) 3ʹ → 5ʹ proofreading activity
A) 5ʹ → 3ʹ polymerase activity
B) joining of Okazaki fragments
C) 5ʹ → 3ʹ exonuclease activity
D) nick translation
E) 3ʹ → 5ʹ proofreading activity

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
38
What is the DNA sequence 5ʹ → 3ʹ) determined in this gel? Assume the samples were applied near the top of the gel. 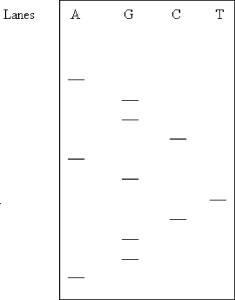
A) AGGCTGGACGA
B) AGGGGCTAGCA
C) AGGCTGACGGA
D) AGGCAGTCGGA

A) AGGCTGGACGA
B) AGGGGCTAGCA
C) AGGCTGACGGA
D) AGGCAGTCGGA

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
39
In packaging the eukaryotic DNA into nucleosomes, it is thought that the requirement for and their synthesis is a cause of the slower rate of movement of the replication fork.
A) Okazaki fragments
B) larger DNA molecules
C) increase number of accessory proteins
D) histone proteins
E) lagging strands
A) Okazaki fragments
B) larger DNA molecules
C) increase number of accessory proteins
D) histone proteins
E) lagging strands

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
40
What is a reasonable length for an RNA primer in E. coli?
A) a few nucleotides
B) 50 nucleotides
C) 100 nucleotides
D) 1000 nucleotides
A) a few nucleotides
B) 50 nucleotides
C) 100 nucleotides
D) 1000 nucleotides

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
41
DNA polymerase III is the largest DNA polymerase in E. coli.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
42
In the Holliday model describing recombination
A) homologous sequences line up.
B) strands of DNA are nicked.
C) DNA strands associate with the homologous strand ʺstrand invasionʺ).
D) strands rotate then are cleaved at the crossover point.
E) All of the above
A) homologous sequences line up.
B) strands of DNA are nicked.
C) DNA strands associate with the homologous strand ʺstrand invasionʺ).
D) strands rotate then are cleaved at the crossover point.
E) All of the above

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
43
Deaminated bases must be removed from DNA because
A) they distort the molecule.
B) they form thymine dimers.
C) they pair incorrectly during replication.
D) the mRNA formed may be irregular.
A) they distort the molecule.
B) they form thymine dimers.
C) they pair incorrectly during replication.
D) the mRNA formed may be irregular.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
44
The purpose of SSB is to prevent single-stranded DNA from folding back on itself to form double-helical regions.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
45
During recombination in E. coli, recognizes) regions of sequence similarity and promotes) strand invasion, while binds) to a Holliday junction promoting strand migration.
A) RuvA and RuvB; RecBCD
B) RecBCD; RecA
C) RecA; RecBCD
D) RecA; RuvA and RuvB
A) RuvA and RuvB; RecBCD
B) RecBCD; RecA
C) RecA; RecBCD
D) RecA; RuvA and RuvB

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
46
The general excision repair pathway for DNA repair has the following order:
A) endonuclease cuts → helicase or endonuclease removes → DNA polymerase → DNA ligase
B) DNA ligase → DNA polymerase → helicase or endonuclease removes → endonuclease cuts
C) endonuclease cuts → DNA polymerase repairs → helicase opens up → DNA ligase
D) endonuclease cuts → DNA ligase removes → endonuclease removes → DNA ligase
A) endonuclease cuts → helicase or endonuclease removes → DNA polymerase → DNA ligase
B) DNA ligase → DNA polymerase → helicase or endonuclease removes → endonuclease cuts
C) endonuclease cuts → DNA polymerase repairs → helicase opens up → DNA ligase
D) endonuclease cuts → DNA ligase removes → endonuclease removes → DNA ligase

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
47
Proteins bind to both origin sites and termination sites on DNA during DNA replication.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
48
The sequence of proteins used in bacterial recombination is
A) RuvC → RuvB → RecA → RecBCD
B) RecBCD → RecA → RuvAB → RuvC
C) RecA → RecBCD → RuvAB → RuvC
D) RuvAB → RuvC → RecBCD → RecA
A) RuvC → RuvB → RecA → RecBCD
B) RecBCD → RecA → RuvAB → RuvC
C) RecA → RecBCD → RuvAB → RuvC
D) RuvAB → RuvC → RecBCD → RecA

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
49
Why is DNA damage in skin cells from exposure to excessive UV light not completely reversed by photoreactivation?
A) DNA photolyase in humans is activated only under conditions of heat shock.
B) Humans lack a photoreactivation repair mechanism.
C) The presence of histones greatly slows photoreactivation, thus humans depends primarily on other forms of DNA repair.
D) Photoreactivation is specific for the removal of thymine dimers. Thymine dimers do not form in human DNA.
A) DNA photolyase in humans is activated only under conditions of heat shock.
B) Humans lack a photoreactivation repair mechanism.
C) The presence of histones greatly slows photoreactivation, thus humans depends primarily on other forms of DNA repair.
D) Photoreactivation is specific for the removal of thymine dimers. Thymine dimers do not form in human DNA.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
50
In the Sanger method for DNA sequencing, the gels are read from bottom to top, since the smallest fragments travel farthest in the sequencing gel.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
51
The β subunits of E. coli DNA polymerase form a sliding clamp that surrounds the DNA strands at the replication fork.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
52
The polymerization of DNA is essentially irreversible due to the hydrolysis of pyrophosphate.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
53
How are the BRCA proteins and their genes implicated in breast cancer?
A) The BRCA proteins damage DNA by generating double-stranded breaks which can lead to cancer.
B) Cells become more susceptible to damage if the genes for these proteins are damaged.
C) BRCA proteins inhibit excision repair mechanisms.
D) BRCA proteins initiate recombination between non-homologous DNA sequences.
A) The BRCA proteins damage DNA by generating double-stranded breaks which can lead to cancer.
B) Cells become more susceptible to damage if the genes for these proteins are damaged.
C) BRCA proteins inhibit excision repair mechanisms.
D) BRCA proteins initiate recombination between non-homologous DNA sequences.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
54
DNA polymerase I possesses 5ʹ → 3ʹ exonuclease activity.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
55
The chromosome of the fruit fly D. melanogaster is about twice as large as the E. coli
chromosome.
chromosome.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
56
In DNA repair, DNA glycosylases recognize deaminated bases within the DNA polymer and remove the deaminated
A) nucleoside.
B) nucleotide.
C) thymine dimers.
D) bases.
A) nucleoside.
B) nucleotide.
C) thymine dimers.
D) bases.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
57
On average about twenty errors are made during each replication of the human genome.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
58
All DNA ligase enzymes use ATP as a cosubstrate.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
59
In E. coli replication begins at the origin of replication and proceeds in one direction until the entire circular DNA molecule has been copied.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
60
Prokaryotic DNA replication occurs in two steps. First, ATP provides a phosphate to the growing DNA chain. This is followed by addition of a nucleoside.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
61
Sanger DNA sequencing requires inhibition of DNA polymerization by a specific dideoxynucleoside every time it is copied in the replication process.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
62
A short primer is added to the DNA polymerase reaction mix for Sanger DNA sequencing that has a 5ʹ free hydroxyl to which new nucleotides can be added.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
63
Eukaryotic cells contain at least five DNA polymerases all of which are responsible for nuclear DNA replication.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
64
There are more replication forks during replication of eukaryotic DNA than prokaryotic DNA.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
65
Since the rate of form movement in eukaryotes is much slower than that of bacteria, chromosomes require more than an hour to replicate.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
66
The Klenow fragment can be used for DNA sequencing if proteins such as SSB protein are added.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
67
DNA polymerase I which is inhibited by dideoxynucleoside triphosphates is used in the Sanger method for DNA sequencing.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
68
DNA repair cannot be done without breaking the phosphodiester backbone of DNA.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
69
All recombination events involve homologous recombination between closely related DNA sequences.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
70
At one point during DNA recombination in E. coli there is formation of a triple-stranded DNA.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
71
Although histones and DNA are made in the same parts of the eukaryotic cell, the packaging of them in the nucleosome slows down replication so that it is slower than that of prokaryotes.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
72
In a sequencing gel, all fragment in one of the four lanes terminates in the same type of nucleotide base.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
73
Since deamination is a common cause of DNA damage, and since the deamination of cytosine forms a product similar in structure to uracil, repair enzymes cannot repair this damage easily.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
74
Homologous recombination is the exchange of DNA between closely related DNA sequences.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
75
DNA is the only cell macromolecule that can be repaired.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck
76
DNA fingerprinting is possible for identification because each person accumulates over a hundred mutations during his or her lifetime.

Unlock Deck
Unlock for access to all 76 flashcards in this deck.
Unlock Deck
k this deck


