Deck 30: DNA Replication, Repair, and Recombination
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/20
Play
Full screen (f)
Deck 30: DNA Replication, Repair, and Recombination
1
Translating nucleotide sequences into amino acid sequences
(Integrates with Chapter 12.) The following sequence represents part of the nucleotide sequence of a cloned cDNA:
... CAATACGAAGCAATCCCGCGACTAGACCTTAAC...
Can you reach an unambiguous conclusion from these data about the partial amino acid sequence of the protein encoded by this cDNA
(Integrates with Chapter 12.) The following sequence represents part of the nucleotide sequence of a cloned cDNA:
... CAATACGAAGCAATCCCGCGACTAGACCTTAAC...
Can you reach an unambiguous conclusion from these data about the partial amino acid sequence of the protein encoded by this cDNA
To make any conclusions about the cDNA strand we must first examine the sequence. After converting to its RNA form, we get the following strand.
CAAUACGAAGCAAUCCCGCGACUAGACCUUAAC
First thing is to determine a reading frame by looking for a start codon, AUG. There are no start codons in the sequence.
Next we look for stop codons: UGA, UAG, or UAA.
CAAUACGAAGCAAUCCCGCGAC UAG ACCU UAA C
We now can see that there are stop codons for 2 of the 3 possible reading frames. This means that the preceding codons, account for the C-terminal of the protein. The 3 rd reading frame would start with CAA and would not have a stop codon in the portion of the DNA sequence that we are provided.
More information would be required to determine which is the correct reading frame.
CAAUACGAAGCAAUCCCGCGACUAGACCUUAAC
First thing is to determine a reading frame by looking for a start codon, AUG. There are no start codons in the sequence.
Next we look for stop codons: UGA, UAG, or UAA.
CAAUACGAAGCAAUCCCGCGAC UAG ACCU UAA C
We now can see that there are stop codons for 2 of the 3 possible reading frames. This means that the preceding codons, account for the C-terminal of the protein. The 3 rd reading frame would start with CAA and would not have a stop codon in the portion of the DNA sequence that we are provided.
More information would be required to determine which is the correct reading frame.
2
Nucleotide sequences, possible codons, and amino acid specification
A random (AG) copolymer was synthesized with polynucleotide phosphorylase using a mixture of 5 parts adenine nucleotide to one part guanine nucleotide as substrate. If this random copolymer is used in a cell-free protein synthesis system, which amino acids will be incorporated into the polypeptide product What will be the relative abundances of these amino acids in the product
A random (AG) copolymer was synthesized with polynucleotide phosphorylase using a mixture of 5 parts adenine nucleotide to one part guanine nucleotide as substrate. If this random copolymer is used in a cell-free protein synthesis system, which amino acids will be incorporated into the polypeptide product What will be the relative abundances of these amino acids in the product
The following in vitro reaction is catalyzed by polynucleotide phosphorylase:
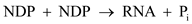 In this instance NDP accounts for 5 parts ADP for every 1 part GDP. This means that the probability that a given NDP will be ADP is 5/6 and the probability that a given NDP will be GDP is 1/6.
In this instance NDP accounts for 5 parts ADP for every 1 part GDP. This means that the probability that a given NDP will be ADP is 5/6 and the probability that a given NDP will be GDP is 1/6.
There are 2 3 or 8 possible codon combinations that can be produced from a mixture of the 2 nucleotides. The probability of creating a specific codon can be calculated by use of the product rule. For example, for the codon GGG, the probability is
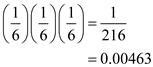 The 8 possible codons for a mixture of A and G is:
The 8 possible codons for a mixture of A and G is:
AAA-Lys
AAG-Lys
AGA-Arg
GAA-Glu
AGG-Arg
GAG-Glu
GGA-Gly
GGG-Gly
The following chart shows the probability of each codon and the amino acid that it codes for.
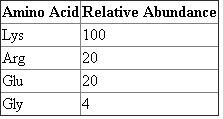
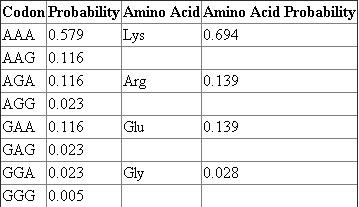 The relative abundance of each of these amino acids in the product can be determined by dividing the individual amino acid probabilities by the probability of the most probable amino acid, which in this case is Lys, and multiplying by 100.
The relative abundance of each of these amino acids in the product can be determined by dividing the individual amino acid probabilities by the probability of the most probable amino acid, which in this case is Lys, and multiplying by 100.
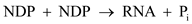 In this instance NDP accounts for 5 parts ADP for every 1 part GDP. This means that the probability that a given NDP will be ADP is 5/6 and the probability that a given NDP will be GDP is 1/6.
In this instance NDP accounts for 5 parts ADP for every 1 part GDP. This means that the probability that a given NDP will be ADP is 5/6 and the probability that a given NDP will be GDP is 1/6.There are 2 3 or 8 possible codon combinations that can be produced from a mixture of the 2 nucleotides. The probability of creating a specific codon can be calculated by use of the product rule. For example, for the codon GGG, the probability is
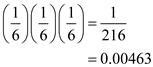 The 8 possible codons for a mixture of A and G is:
The 8 possible codons for a mixture of A and G is:AAA-Lys
AAG-Lys
AGA-Arg
GAA-Glu
AGG-Arg
GAG-Glu
GGA-Gly
GGG-Gly
The following chart shows the probability of each codon and the amino acid that it codes for.
 The relative abundance of each of these amino acids in the product can be determined by dividing the individual amino acid probabilities by the probability of the most probable amino acid, which in this case is Lys, and multiplying by 100.
The relative abundance of each of these amino acids in the product can be determined by dividing the individual amino acid probabilities by the probability of the most probable amino acid, which in this case is Lys, and multiplying by 100.
3
The second genetic code
Review the evidence establishing that aminoacyl-tRNA synthetases bridge the information gap between amino acids and codons. Indicate the various levels of specificity possessed by aminoacyltRNA synthetases that are essential for high-fidelity translation of messenger RNA molecules.
Review the evidence establishing that aminoacyl-tRNA synthetases bridge the information gap between amino acids and codons. Indicate the various levels of specificity possessed by aminoacyltRNA synthetases that are essential for high-fidelity translation of messenger RNA molecules.
Francis Crick's central dogmas of genetics states that DNA is transcribed into RNA and RNA is translated into proteins. The formation of the correct protein is strongly dependent on the ability of tRNA to carry the correct amino acid.
Aminoacyl-tRNA synthetases are the enzyme that charges the tRNA with an amino acid in a 2 step process. First the amino acid is adenylated to form aminoacyl-adenylate. This process is dependent on the correct amino acid sequence. Errors in this process do occur, but at low frequencies. An error at this stage is referred to as misacylation. When this occurs the Aminoacyl tRNA synthetases will not release the amino acid and a deacylase will be activated to hydrolyze the tRNA. The aminoacyl-adenylate is then transferred to the 2'-OH or 3'-OH of tRNA.
Aminoacyl-tRNA synthetases are the enzyme that charges the tRNA with an amino acid in a 2 step process. First the amino acid is adenylated to form aminoacyl-adenylate. This process is dependent on the correct amino acid sequence. Errors in this process do occur, but at low frequencies. An error at this stage is referred to as misacylation. When this occurs the Aminoacyl tRNA synthetases will not release the amino acid and a deacylase will be activated to hydrolyze the tRNA. The aminoacyl-adenylate is then transferred to the 2'-OH or 3'-OH of tRNA.
4
Codon-anticodon recognition: base-pairing possibilities
(Integrates with Chapter 11.) Draw base-pair structures for (a) a G:C base pair, (b) a C:G base pair, (c) a G:U base pair, and (d) a U:G base pair. Note how these various base pairs differ in the potential hydrogen-bonding patterns they present within the major groove and minor groove of a double-helical nucleic acid.
(Integrates with Chapter 11.) Draw base-pair structures for (a) a G:C base pair, (b) a C:G base pair, (c) a G:U base pair, and (d) a U:G base pair. Note how these various base pairs differ in the potential hydrogen-bonding patterns they present within the major groove and minor groove of a double-helical nucleic acid.

Unlock Deck
Unlock for access to all 20 flashcards in this deck.
Unlock Deck
k this deck
5
Consequences of the wobble hypothesis
Point out why Crick's wobble hypothesis would allow fewer than 61 anticodons to be used to translate the 61 sense codons. How does "wobble" tend to accelerate the rate of translation
Point out why Crick's wobble hypothesis would allow fewer than 61 anticodons to be used to translate the 61 sense codons. How does "wobble" tend to accelerate the rate of translation

Unlock Deck
Unlock for access to all 20 flashcards in this deck.
Unlock Deck
k this deck
6
Sense-to-nonsense codon mutations
How many codons can mutate to become nonsense codons through a single base change Which amino acids do they encode
How many codons can mutate to become nonsense codons through a single base change Which amino acids do they encode

Unlock Deck
Unlock for access to all 20 flashcards in this deck.
Unlock Deck
k this deck
7
Amino acid possibilities in nonsense suppression
Nonsense suppression occurs when a suppressor mutant arises that reads a nonsense codon and inserts an amino acid, as if the nonsense codon was actually a sense codon. Which amino acids do you think are most likely to be incorporated by nonsense suppressor mutants
Nonsense suppression occurs when a suppressor mutant arises that reads a nonsense codon and inserts an amino acid, as if the nonsense codon was actually a sense codon. Which amino acids do you think are most likely to be incorporated by nonsense suppressor mutants

Unlock Deck
Unlock for access to all 20 flashcards in this deck.
Unlock Deck
k this deck
8
Prokaryotic vs. eukaryotic protein synthesis
Why do you suppose eukaryotic protein synthesis is only 10% as fast as prokaryotic protein synthesis
Why do you suppose eukaryotic protein synthesis is only 10% as fast as prokaryotic protein synthesis

Unlock Deck
Unlock for access to all 20 flashcards in this deck.
Unlock Deck
k this deck
9
Capacity of the ribosomal peptide exit tunnel
If the tunnel through the large ribosomal subunit is 10 nm long, how many amino acid residues might be contained within it (Assume that the growing polypeptide chain is in an extended -sheet- like conformation.)
If the tunnel through the large ribosomal subunit is 10 nm long, how many amino acid residues might be contained within it (Assume that the growing polypeptide chain is in an extended -sheet- like conformation.)

Unlock Deck
Unlock for access to all 20 flashcards in this deck.
Unlock Deck
k this deck
10
The consequences of ribosome complexity
Eukaryotic ribosomes are larger and more complex than prokaryotic ribosomes. What advantages and disadvantages might this greater ribosomal complexity bring to a eukaryotic cell
Eukaryotic ribosomes are larger and more complex than prokaryotic ribosomes. What advantages and disadvantages might this greater ribosomal complexity bring to a eukaryotic cell

Unlock Deck
Unlock for access to all 20 flashcards in this deck.
Unlock Deck
k this deck
11
Ribosomes as two-subunit structures
What ideas can you suggest to explain why ribosomes invariably exist as two-subunit structures, instead of a larger, single-subunit entity
What ideas can you suggest to explain why ribosomes invariably exist as two-subunit structures, instead of a larger, single-subunit entity

Unlock Deck
Unlock for access to all 20 flashcards in this deck.
Unlock Deck
k this deck
12
tRNAi Met recognition and translation initiation
How do prokaryotic cells determine whether a particular methionyl-tRNA Met is intended to initiate protein synthesis or to deliver a Met residue for internal incorporation into a polypeptide chain How do the Met codons for these two different purposes differ How do eukaryotic cells handle these problems
How do prokaryotic cells determine whether a particular methionyl-tRNA Met is intended to initiate protein synthesis or to deliver a Met residue for internal incorporation into a polypeptide chain How do the Met codons for these two different purposes differ How do eukaryotic cells handle these problems

Unlock Deck
Unlock for access to all 20 flashcards in this deck.
Unlock Deck
k this deck
13
The function of the Shine-Dalgarno sequence
What is the Shine-Dalgarno sequence What does it do The efficiency of protein synthesis initiation may vary by as much as 100-fold for different mRNAs. How might the Shine-Dalgarno sequence be responsible for this difference
What is the Shine-Dalgarno sequence What does it do The efficiency of protein synthesis initiation may vary by as much as 100-fold for different mRNAs. How might the Shine-Dalgarno sequence be responsible for this difference

Unlock Deck
Unlock for access to all 20 flashcards in this deck.
Unlock Deck
k this deck
14
Reconciling peptide bond formation and tRNA movements
In the protein synthesis elongation events described under the section on translocation, which of the following seems the most apt account of the peptidyl transfer reaction: (a) The peptidyl-tRNA delivers its peptide chain to the newly arrived aminoacyl-tRNA situated in the A site, or (b) the aminoacyl end of the aminoacyl-tRNA moves toward the P site to accept the peptidyl chain Which of these two scenarios makes most sense to you Why
In the protein synthesis elongation events described under the section on translocation, which of the following seems the most apt account of the peptidyl transfer reaction: (a) The peptidyl-tRNA delivers its peptide chain to the newly arrived aminoacyl-tRNA situated in the A site, or (b) the aminoacyl end of the aminoacyl-tRNA moves toward the P site to accept the peptidyl chain Which of these two scenarios makes most sense to you Why

Unlock Deck
Unlock for access to all 20 flashcards in this deck.
Unlock Deck
k this deck
15
Why translation factors are G proteins
(Integrates with Chapter 15.) Why might you suspect that the elongation factors EF-Tu and EF-Ts are evolutionarily related to the G proteins of membrane signal transduction pathways described in Chapter 15
(Integrates with Chapter 15.) Why might you suspect that the elongation factors EF-Tu and EF-Ts are evolutionarily related to the G proteins of membrane signal transduction pathways described in Chapter 15

Unlock Deck
Unlock for access to all 20 flashcards in this deck.
Unlock Deck
k this deck
16
The energetic cost of peptide elongation
How many ATP equivalents are consumed for each amino acid added to an elongating polypeptide chain during the process of protein synthesis
How many ATP equivalents are consumed for each amino acid added to an elongating polypeptide chain during the process of protein synthesis

Unlock Deck
Unlock for access to all 20 flashcards in this deck.
Unlock Deck
k this deck
17
Exploring the structure of the 30S ribosomal subunit
Go to www.pdb.org and bring up PDB file 1GIX, which shows the 30S ribosomal subunit, the three tRNAs, and mRNA. In the box on the right titled "Biological Assembly", click "More Images…", and then scroll down to look at the Interactive View. By moving your cursor over the image, you can rotate it to view it from any perspective.
a. How are the ribosomal proteins represented in the image
b. How is the 16S rRNA portrayed
c. Rotate the image to see how the tRNAs stick out from the structure. Which end of the tRNA is sticking out
d. Where will these ends of the tRNAs lie when the 50S subunit binds to this complex
Go to www.pdb.org and bring up PDB file 1GIX, which shows the 30S ribosomal subunit, the three tRNAs, and mRNA. In the box on the right titled "Biological Assembly", click "More Images…", and then scroll down to look at the Interactive View. By moving your cursor over the image, you can rotate it to view it from any perspective.
a. How are the ribosomal proteins represented in the image
b. How is the 16S rRNA portrayed
c. Rotate the image to see how the tRNAs stick out from the structure. Which end of the tRNA is sticking out
d. Where will these ends of the tRNAs lie when the 50S subunit binds to this complex

Unlock Deck
Unlock for access to all 20 flashcards in this deck.
Unlock Deck
k this deck
18
Exploring the structure of the 50S ribosomal subunit
Go back to www.pdb.org and bring up PDB file 1FFK, which shows the 50S ribosomal subunit. In the box titled "Biological Assembly," click "More Images," and scroll down to look at the Interactive View. Right-click the image to discover more information and tools.
a. How many atoms are represented in this structure
b. Are the bases of the nucleotides visible anywhere in the structure
c. Can you find double helical regions of RNA
d. Right-click and, under "Select," select all proteins. Right-click again and select "Render," then "Scheme," and then "CPK Spacefill" to highlight the ribosomal proteins. Go back and cancel the protein selection. Then select "Nucleic," and render nucleic acid in "CPK Spacefill." Which macromolecular species seems to predominate the structure
Go back to www.pdb.org and bring up PDB file 1FFK, which shows the 50S ribosomal subunit. In the box titled "Biological Assembly," click "More Images," and scroll down to look at the Interactive View. Right-click the image to discover more information and tools.
a. How many atoms are represented in this structure
b. Are the bases of the nucleotides visible anywhere in the structure
c. Can you find double helical regions of RNA
d. Right-click and, under "Select," select all proteins. Right-click again and select "Render," then "Scheme," and then "CPK Spacefill" to highlight the ribosomal proteins. Go back and cancel the protein selection. Then select "Nucleic," and render nucleic acid in "CPK Spacefill." Which macromolecular species seems to predominate the structure

Unlock Deck
Unlock for access to all 20 flashcards in this deck.
Unlock Deck
k this deck
19
Review the list of Shine-Dalgarno sequences in Figure 30.18 and select the one that will interact best with the 3'-end of E. coli 16S rRNA.

Unlock Deck
Unlock for access to all 20 flashcards in this deck.
Unlock Deck
k this deck
20
Chloramphenicol (Figure 30.31) inhibits the peptidyl transferase activity of the 50S ribosomal subunit. The 50S peptidyl transferase active site consists solely of functionalities provided by the 23S rRNA. What sorts of interactions do you think take place when chloramphenicol binds to the peptidyl transferase center Which groups on chloramphenicol might be involved in these interactions

Unlock Deck
Unlock for access to all 20 flashcards in this deck.
Unlock Deck
k this deck


