Deck 28: Nucleotide Metabolism
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/19
Play
Full screen (f)
Deck 28: Nucleotide Metabolism
1
Semiconservative or conservative DNA replication
If 15 N-labeled E. coli DNA has a density of 1.724 g/mL, 14 N-labeled DNA has a density of 1.710 g/mL, and E. coli cells grown for many generations on 14 NH 4 + as nitrogen source are transferred to media containing 15 NH 4 + as sole N-source, (a) what will be the density of the DNA after one generation, assuming replication is semiconservative (b) Suppose replication took place by a conservative mechanism in which the parental strands remained together and the two progeny strands were paired. Design an experiment that could distinguish between semiconservative and conservative modes of replication.
If 15 N-labeled E. coli DNA has a density of 1.724 g/mL, 14 N-labeled DNA has a density of 1.710 g/mL, and E. coli cells grown for many generations on 14 NH 4 + as nitrogen source are transferred to media containing 15 NH 4 + as sole N-source, (a) what will be the density of the DNA after one generation, assuming replication is semiconservative (b) Suppose replication took place by a conservative mechanism in which the parental strands remained together and the two progeny strands were paired. Design an experiment that could distinguish between semiconservative and conservative modes of replication.
(a) The density for both of the DNA strands after one generation in a semi-conservative model can be calculated by finding the average density of the heavy and light strands.
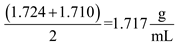 (b) To distinguish between the semi conservative and conservative models, you would first denature the strand being tested intO2 single strands and then analyze it via CsCl density gradient centrifugation. In a conservative model, you would expect the densities of the 2 strands to be the same. In a semi-conservative model you would expect the 2 strands to have different densities. This would be because one will be 15 N DNA and the other one will be 14 N DNA.
(b) To distinguish between the semi conservative and conservative models, you would first denature the strand being tested intO2 single strands and then analyze it via CsCl density gradient centrifugation. In a conservative model, you would expect the densities of the 2 strands to be the same. In a semi-conservative model you would expect the 2 strands to have different densities. This would be because one will be 15 N DNA and the other one will be 14 N DNA.
After a round of replication, we would see 2 bands for conservative replication. One is the DNA with density relating to 15 N DNA and the other one is DNA with 14 N DNA. If there are any additional rounds of replication, the amount of 15 N DNA will increase, while the amount of 14 N DNA will remain constant.
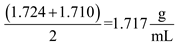 (b) To distinguish between the semi conservative and conservative models, you would first denature the strand being tested intO2 single strands and then analyze it via CsCl density gradient centrifugation. In a conservative model, you would expect the densities of the 2 strands to be the same. In a semi-conservative model you would expect the 2 strands to have different densities. This would be because one will be 15 N DNA and the other one will be 14 N DNA.
(b) To distinguish between the semi conservative and conservative models, you would first denature the strand being tested intO2 single strands and then analyze it via CsCl density gradient centrifugation. In a conservative model, you would expect the densities of the 2 strands to be the same. In a semi-conservative model you would expect the 2 strands to have different densities. This would be because one will be 15 N DNA and the other one will be 14 N DNA.After a round of replication, we would see 2 bands for conservative replication. One is the DNA with density relating to 15 N DNA and the other one is DNA with 14 N DNA. If there are any additional rounds of replication, the amount of 15 N DNA will increase, while the amount of 14 N DNA will remain constant.
2
The enzymatic activities of DNA polymerase I
(a) What are the respective roles of the 5'-exonuclease and 3'-exonuclease activities of DNA polymerase I (b) What would be a feature of an E. coli strain that lacked DNA polymerase I 3'- exonuclease activity
(a) What are the respective roles of the 5'-exonuclease and 3'-exonuclease activities of DNA polymerase I (b) What would be a feature of an E. coli strain that lacked DNA polymerase I 3'- exonuclease activity
The importance of DNA polymerase functions in replaces RNA primers with DNA during replication and fill gaps produced by DNA repair endonucleases during DNA repair. The role of 5 ' -exonulease activity is to remove DNA primers. In the other hand the 3 ' -exonuclease activity is an editor. Polymerase uses its 5 ' -exonuclease activity to remove mononucleotides and oligonucleotides.
3
Multiple replication forks in E. coli I
Assuming DNA replication proceeds at a rate of 750 base pairs per second, calculate how long it will take to replicate the entire E. coli genome. Under optimal conditions, E. coli cells divide every 20 minutes. What is the minimal number of replication forks per E. coli chromosome in order to sustain such a rate of cell division
Assuming DNA replication proceeds at a rate of 750 base pairs per second, calculate how long it will take to replicate the entire E. coli genome. Under optimal conditions, E. coli cells divide every 20 minutes. What is the minimal number of replication forks per E. coli chromosome in order to sustain such a rate of cell division
Knowing that the E. coli genome is 4.64
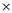 10 6 bp, we predict that DNA replication proceeds at a rate of 750 bp per second. The time required to replicate this amount of DNA is
10 6 bp, we predict that DNA replication proceeds at a rate of 750 bp per second. The time required to replicate this amount of DNA is
 We can determine that rate of replication in order for the genome to be replicated in 20 minutes.
We can determine that rate of replication in order for the genome to be replicated in 20 minutes.
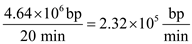 At the bidirectional origin of replication, DNA is being synthesized at the rate of
At the bidirectional origin of replication, DNA is being synthesized at the rate of
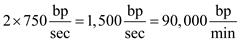 Then we can determine how many replication bubbles exist
Then we can determine how many replication bubbles exist
 If there are 2.58 replication bubbles, there are 5.16 replication forks.
If there are 2.58 replication bubbles, there are 5.16 replication forks.
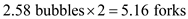
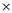 10 6 bp, we predict that DNA replication proceeds at a rate of 750 bp per second. The time required to replicate this amount of DNA is
10 6 bp, we predict that DNA replication proceeds at a rate of 750 bp per second. The time required to replicate this amount of DNA is We can determine that rate of replication in order for the genome to be replicated in 20 minutes.
We can determine that rate of replication in order for the genome to be replicated in 20 minutes.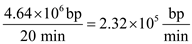 At the bidirectional origin of replication, DNA is being synthesized at the rate of
At the bidirectional origin of replication, DNA is being synthesized at the rate of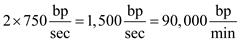 Then we can determine how many replication bubbles exist
Then we can determine how many replication bubbles exist If there are 2.58 replication bubbles, there are 5.16 replication forks.
If there are 2.58 replication bubbles, there are 5.16 replication forks.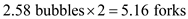
4
Multiple replication forks in E. coli II
On the basis of Figure 28.2, draw a simple diagram illustrating replication of the circular E. coli chromosome (a) at an early stage, (b) when one-third completed, (c) when two-thirds completed, and (d) when almost finished, assuming the initiation of replication at oriC has occurred only once. Then, draw a diagram showing the E. coli chromosome in problem 3 where the E. coli cell is dividing every 20 minutes.
On the basis of Figure 28.2, draw a simple diagram illustrating replication of the circular E. coli chromosome (a) at an early stage, (b) when one-third completed, (c) when two-thirds completed, and (d) when almost finished, assuming the initiation of replication at oriC has occurred only once. Then, draw a diagram showing the E. coli chromosome in problem 3 where the E. coli cell is dividing every 20 minutes.

Unlock Deck
Unlock for access to all 19 flashcards in this deck.
Unlock Deck
k this deck
5
Molecules of DNA polymerase III per cell vs. growth rate
It is estimated that there are forty molecules of DNA polymerase III per E. coli cell. Is it likely that E. coli growth rate is limited by DNA polymerase III availability
It is estimated that there are forty molecules of DNA polymerase III per E. coli cell. Is it likely that E. coli growth rate is limited by DNA polymerase III availability

Unlock Deck
Unlock for access to all 19 flashcards in this deck.
Unlock Deck
k this deck
6
Number of Okazaki fragments in E. coli and human DNA replication
Approximately how many Okazaki fragments are synthesized in the course of replicating an E. coli chromosome How many in replicating an "average" human chromosome
Approximately how many Okazaki fragments are synthesized in the course of replicating an E. coli chromosome How many in replicating an "average" human chromosome

Unlock Deck
Unlock for access to all 19 flashcards in this deck.
Unlock Deck
k this deck
7
The roles of helicases and gyrases
How do DNA gyrases and helicases differ in their respective functions and modes of action
How do DNA gyrases and helicases differ in their respective functions and modes of action

Unlock Deck
Unlock for access to all 19 flashcards in this deck.
Unlock Deck
k this deck
8
Human Genome Replication Rate Assume DNA replication proceeds at a rate of 100 base pairs per second in human cells and origins of replication occur every 300 kbp. Assume also that DNA polymerase III is highly processive and only 2 molecules of DNA polymerase III are needed per replication fork. How long would it take to replicate the entire diploid human genome How many molecules of DNA polymerase does each cell need to carry out this task

Unlock Deck
Unlock for access to all 19 flashcards in this deck.
Unlock Deck
k this deck
9
Heteroduplex DNA formation in recombination
From the information in Figure 28.17 and 28.18, diagram the recombinational event leading to the formation of a heteroduplex DNA region within a bacteriophage chromosome.
From the information in Figure 28.17 and 28.18, diagram the recombinational event leading to the formation of a heteroduplex DNA region within a bacteriophage chromosome.

Unlock Deck
Unlock for access to all 19 flashcards in this deck.
Unlock Deck
k this deck
10
Homologous recombination, heteroduplex DNA, and mismatch repair
Homologous recombination in E. coli leads to the formation of regions of heteroduplex DNA. By definition, such regions contain mismatched bases. Why doesn't the mismatch repair system of E. coli eliminate these mismatches
Homologous recombination in E. coli leads to the formation of regions of heteroduplex DNA. By definition, such regions contain mismatched bases. Why doesn't the mismatch repair system of E. coli eliminate these mismatches

Unlock Deck
Unlock for access to all 19 flashcards in this deck.
Unlock Deck
k this deck
11
RecA:DNA nucleoprotein helix/B-DNA comparison
If RecA protein unwinds duplex DNA so that there are about 18.6 bp per turn, what is the change in the helical twist of DNA, compared to its value in B-DNA
If RecA protein unwinds duplex DNA so that there are about 18.6 bp per turn, what is the change in the helical twist of DNA, compared to its value in B-DNA

Unlock Deck
Unlock for access to all 19 flashcards in this deck.
Unlock Deck
k this deck
12
Chemical mutagenesis of DNA bases
Show the nucleotide sequence changes that might arise in a dsDNA (coding strand segment GCTA) upon mutagenesis with (a) HNO2 , (b) bromouracil, and (c) 2-aminopurine.
Show the nucleotide sequence changes that might arise in a dsDNA (coding strand segment GCTA) upon mutagenesis with (a) HNO2 , (b) bromouracil, and (c) 2-aminopurine.

Unlock Deck
Unlock for access to all 19 flashcards in this deck.
Unlock Deck
k this deck
13
Transposons as mutagens
Transposons are mutagenic agents. Why
Transposons are mutagenic agents. Why

Unlock Deck
Unlock for access to all 19 flashcards in this deck.
Unlock Deck
k this deck
14
Give a plausible explanation for the genetic and infectious properties of
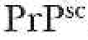
.
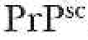
.

Unlock Deck
Unlock for access to all 19 flashcards in this deck.
Unlock Deck
k this deck
15
Helicase unwinding of the E. coli chromosome
Hexameric helicases, such as DnaB, the MCM proteins, and papilloma virus E1 helicase (illustrated in Figures 16.23-16.25), unwind DNA by passing one strand of the DNA duplex through the central pore, using a mechanism based on ATP-dependent binding interactions with the bases of that strand. The genome of E. coli K12 consists of 4,686,137 nucleotides. Assuming that DnaB functions like papilloma virus E1 helicase, from the information given in Chapter 16 on ATP-coupled DNA unwinding, calculate how many molecules of ATP would be needed to completely unwind the E. coli K12 chromosome.
Hexameric helicases, such as DnaB, the MCM proteins, and papilloma virus E1 helicase (illustrated in Figures 16.23-16.25), unwind DNA by passing one strand of the DNA duplex through the central pore, using a mechanism based on ATP-dependent binding interactions with the bases of that strand. The genome of E. coli K12 consists of 4,686,137 nucleotides. Assuming that DnaB functions like papilloma virus E1 helicase, from the information given in Chapter 16 on ATP-coupled DNA unwinding, calculate how many molecules of ATP would be needed to completely unwind the E. coli K12 chromosome.

Unlock Deck
Unlock for access to all 19 flashcards in this deck.
Unlock Deck
k this deck
16
Translesion DNA polymerase IV experiment
Asako Furukohri, Myron F. Goodman, and Hisaji Maki wanted to discover how the translesion DNA polymerase IV takes over from DNA polymerase III at a stalled replication fork (see Journal of Biological Chemistry 283:11260-11269, 2008). They showed that DNA polymerase IV could displace DNA polymerase III from a stalled replication fork formed in an in vitro system containing DNA, DNA polymerase III, the -clamp, and SSB. Devise your own experiment to show how such displacement might be demonstrated. (Hint: Assume that you have protein identification tools that allow you to distinguish easily between DNA polymerase III and DNA polymerase IV.)
Asako Furukohri, Myron F. Goodman, and Hisaji Maki wanted to discover how the translesion DNA polymerase IV takes over from DNA polymerase III at a stalled replication fork (see Journal of Biological Chemistry 283:11260-11269, 2008). They showed that DNA polymerase IV could displace DNA polymerase III from a stalled replication fork formed in an in vitro system containing DNA, DNA polymerase III, the -clamp, and SSB. Devise your own experiment to show how such displacement might be demonstrated. (Hint: Assume that you have protein identification tools that allow you to distinguish easily between DNA polymerase III and DNA polymerase IV.)

Unlock Deck
Unlock for access to all 19 flashcards in this deck.
Unlock Deck
k this deck
17
Functional consequences of Y-family DNA polymerase structure
The eukaryotic translesion DNA polymerases fall into the Y family of DNA polymerases. Structural studies reveal that their fingers and thumb domains are small and stubby (see Figure 28.10). In addition, Y-family polymerase active sites are more open and less constrained where base pairing leads to selection of a dNTP substrate for the polymerase reaction. Discuss the relevance of these structural differences. Would you expect Y-family polymerases to have 3'-exonuclease activity Explain your answer.
The eukaryotic translesion DNA polymerases fall into the Y family of DNA polymerases. Structural studies reveal that their fingers and thumb domains are small and stubby (see Figure 28.10). In addition, Y-family polymerase active sites are more open and less constrained where base pairing leads to selection of a dNTP substrate for the polymerase reaction. Discuss the relevance of these structural differences. Would you expect Y-family polymerases to have 3'-exonuclease activity Explain your answer.

Unlock Deck
Unlock for access to all 19 flashcards in this deck.
Unlock Deck
k this deck
18
Figure 28.11 depicts the eukaryotic cell cycle. Many cell types "exit" the cell cycle and don't divide for prolonged periods, a state termed G0; some, for example, neurons, never divide again.
a. What stage of the cell cycle do you suppose a cell might be in when it exits the cell cycle and enters G0
b. The cell cycle is controlled by checkpoints, cyclins, and CDKs. Describe how biochemical events involving cyclins and CDKs might control passage of a dividing cell through the cell cycle.
a. What stage of the cell cycle do you suppose a cell might be in when it exits the cell cycle and enters G0
b. The cell cycle is controlled by checkpoints, cyclins, and CDKs. Describe how biochemical events involving cyclins and CDKs might control passage of a dividing cell through the cell cycle.

Unlock Deck
Unlock for access to all 19 flashcards in this deck.
Unlock Deck
k this deck
19
Figure 28.39 gives some examples of recombination in IgG codons 95 and 96, as specified by the V and J genes. List the codon possibilities and the amino acids encoded if recombination occurred in codon 97. Which of these possibilities is least desirable

Unlock Deck
Unlock for access to all 19 flashcards in this deck.
Unlock Deck
k this deck


