Deck 25: Dna Restructuring: Repair, Recombination, Rearrangement, Amplification
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/11
Play
Full screen (f)
Deck 25: Dna Restructuring: Repair, Recombination, Rearrangement, Amplification
1
Predict whether a dam methylase deficiency would increase, decrease, or have no effect upon spontaneous mutation rates, and explain the basis for your prediction.
Dam methylase is an abbreviation form of an enzyme deoxyadenosine methylase. This enzyme adds methyl group to adenine in newly synthesized deoxyribonucleic acid (DNA).
DNA methylation stabilizes the DNA. It also helps in cell differentiation and embryonic development. During replication methylated DNA strand temporarily unmethylated. In the two strands of DNA, only one is unmethylated and other remains methylated. This condition is called hemi methylated. The dam methylase quickly methylates the unmethylated DNA strand after replication.
The deficiency of dam methylase leaves the DNA fully unmethylated. The unmethylated DNA has more chances to undergo mutations. Thus, dam methylase deficiency increases the spontaneous mutations.
The dam methylase enzyme increases the mutation rates. This is because the mismatch repair mechanism would repair the mismatched nucleotide which is correct. This will increase the chances of mutation in the DNA (deoxyribonucleic acid) strand.
DNA methylation stabilizes the DNA. It also helps in cell differentiation and embryonic development. During replication methylated DNA strand temporarily unmethylated. In the two strands of DNA, only one is unmethylated and other remains methylated. This condition is called hemi methylated. The dam methylase quickly methylates the unmethylated DNA strand after replication.
The deficiency of dam methylase leaves the DNA fully unmethylated. The unmethylated DNA has more chances to undergo mutations. Thus, dam methylase deficiency increases the spontaneous mutations.
The dam methylase enzyme increases the mutation rates. This is because the mismatch repair mechanism would repair the mismatched nucleotide which is correct. This will increase the chances of mutation in the DNA (deoxyribonucleic acid) strand.
2
For each DNA repair process in column I, list all characteristics from column II that correctly describe that process. 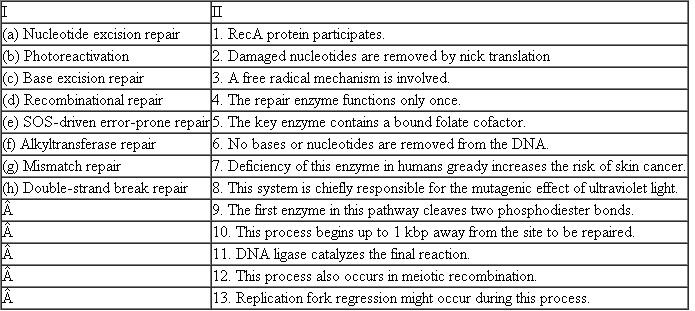

The deoxyribonucleic acid (DNA) is damaged by different sources like ultraviolet (UV) radiations, chemical exposure etc. The damage of DNA is sometimes lethal. Hence, organisms try to repair their DNA by different techniques.
The repair process along with the points that can describe the specific process is as follows:
(a) Nucleotide excision repair: it repairs the DNA damaged by UV radiations. UV damage results in thymine dimers and 6,4-photoproducts. In this mechanism a short single-strand of DNA that contains lesion is removed by nick translation. Basing on the undamaged strand, the DNA polymerase adds nucleotides to the damaged strand. Finally ligation is done by DNA ligase.
Hence, the correct options are 2, 7, 9, and 11.
(b) Photoreactivation: it repairs DNA damaged by exposure to UV radiations. This technique requires visible light for damage repair. This technique has no role in humans but it is best technique in plants. Photolyase enzyme binds to DNA strand and cleaves certain thymine or cytosine dimers which exist in same strand. These dimers bulge the strand thus lesions are formed. The photolyase is a flavoprotein which contains two light-harvesting cofactors. The enzyme acts by electron transfer.
Hence, the correct options are 3, 5, and 6
(c) Base excision repair: it repairs damaged DNA throughout the cell cycle. This mechanism is initiated by DNA glycosylases which recognizes the damaged bases and forms apurinic or apyrimidinic (AP) sites. These sites are cleaved by AP endonuclease. DNA polymerase adds either single nucleotide or short strand of nucleotides to damaged strand. Finally ligation is done by DNA ligase.
Hence, the correct options are 2 and 11.
(d) Recombinational repair: the UV damaged DNA is repaired. It is a complicated process. This technique requires two DNA complexes and exchanges strand from one duplex to another duplex strand. RecA proteins are involved in this process. Finally ligation is done by DNA ligase.
Hence the correct options are 1 and 11.
(e) SOS-driven error-prone repair: in this technique prokaryotic organisms tries to repair its highly damaged DNA at any cost. SOS response is controlled by two proteins called RecA and LexA. In normal cells this technique is turned off. Finally ligation is done by DNA ligase.
Hence the correct options are 1, 8, and 11.
(f) Alkyltransferase repair: the alkylated DNA is repaired by alkyl tranferases. The alkylated base is flipped out of the helix. This is followed by nucleotide excision repair with the recognition of lesions on damaged DNa.Hence the correct options are 4 and 6.
(g) Mismatch repair: this technique is strand specific. During replication, the newly synthesized strand shows an error rate. Hence, in this technique, the newly synthesized DNA strand is repaired. Hemimethylation distinguishes template from newly synthesized strand. The proteins that mediate repair process bind 1 Kb away from site of action and remove the misplaced nucleotide. DNA polymerase adds correct nucleotide to damaged strand. Finally ligation is done by DNA ligase.
Hence the correct options are 10 and 11.
(h) Double strand break repair: it is common in eukaryotic cells. The DNA breaks are joined by action of three enzymes nuclease, polymerase and ligase. This process occurs in meiosis process.
Hence the correct options are 6, 11, and 12.
The repair process along with the points that can describe the specific process is as follows:
(a) Nucleotide excision repair: it repairs the DNA damaged by UV radiations. UV damage results in thymine dimers and 6,4-photoproducts. In this mechanism a short single-strand of DNA that contains lesion is removed by nick translation. Basing on the undamaged strand, the DNA polymerase adds nucleotides to the damaged strand. Finally ligation is done by DNA ligase.
Hence, the correct options are 2, 7, 9, and 11.
(b) Photoreactivation: it repairs DNA damaged by exposure to UV radiations. This technique requires visible light for damage repair. This technique has no role in humans but it is best technique in plants. Photolyase enzyme binds to DNA strand and cleaves certain thymine or cytosine dimers which exist in same strand. These dimers bulge the strand thus lesions are formed. The photolyase is a flavoprotein which contains two light-harvesting cofactors. The enzyme acts by electron transfer.
Hence, the correct options are 3, 5, and 6
(c) Base excision repair: it repairs damaged DNA throughout the cell cycle. This mechanism is initiated by DNA glycosylases which recognizes the damaged bases and forms apurinic or apyrimidinic (AP) sites. These sites are cleaved by AP endonuclease. DNA polymerase adds either single nucleotide or short strand of nucleotides to damaged strand. Finally ligation is done by DNA ligase.
Hence, the correct options are 2 and 11.
(d) Recombinational repair: the UV damaged DNA is repaired. It is a complicated process. This technique requires two DNA complexes and exchanges strand from one duplex to another duplex strand. RecA proteins are involved in this process. Finally ligation is done by DNA ligase.
Hence the correct options are 1 and 11.
(e) SOS-driven error-prone repair: in this technique prokaryotic organisms tries to repair its highly damaged DNA at any cost. SOS response is controlled by two proteins called RecA and LexA. In normal cells this technique is turned off. Finally ligation is done by DNA ligase.
Hence the correct options are 1, 8, and 11.
(f) Alkyltransferase repair: the alkylated DNA is repaired by alkyl tranferases. The alkylated base is flipped out of the helix. This is followed by nucleotide excision repair with the recognition of lesions on damaged DNa.Hence the correct options are 4 and 6.
(g) Mismatch repair: this technique is strand specific. During replication, the newly synthesized strand shows an error rate. Hence, in this technique, the newly synthesized DNA strand is repaired. Hemimethylation distinguishes template from newly synthesized strand. The proteins that mediate repair process bind 1 Kb away from site of action and remove the misplaced nucleotide. DNA polymerase adds correct nucleotide to damaged strand. Finally ligation is done by DNA ligase.
Hence the correct options are 10 and 11.
(h) Double strand break repair: it is common in eukaryotic cells. The DNA breaks are joined by action of three enzymes nuclease, polymerase and ligase. This process occurs in meiosis process.
Hence the correct options are 6, 11, and 12.
3
A 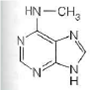
B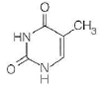
C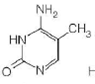
D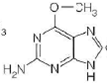
For each of the following characteristics, list all of the bases (A, B, C, or D) to which they apply.
(a) A signal that identifies a parental DNA strand in the MutH,L,S mismatch correction system
(b) Most likely to be involved in cyclobutane dimer formation after ultraviolet irradiation of DNA
(c) A methylated base found immediately to the 5 side of dGMP residues in eukaryotic DNA
(d) Created by treating DNA with alkylating agents that transfer methyl groups and repaired by an "enzyme" that functions only once in its lifetime
(e) Created by AdoMet-dependent methylation of a nucleotide residue in DNA
(f) A substrate for deamination at the DNA level, which would lead to a GC AT transition

B

C

D

For each of the following characteristics, list all of the bases (A, B, C, or D) to which they apply.
(a) A signal that identifies a parental DNA strand in the MutH,L,S mismatch correction system
(b) Most likely to be involved in cyclobutane dimer formation after ultraviolet irradiation of DNA
(c) A methylated base found immediately to the 5 side of dGMP residues in eukaryotic DNA
(d) Created by treating DNA with alkylating agents that transfer methyl groups and repaired by an "enzyme" that functions only once in its lifetime
(e) Created by AdoMet-dependent methylation of a nucleotide residue in DNA
(f) A substrate for deamination at the DNA level, which would lead to a GC AT transition
The structures given in the problem are as follows: 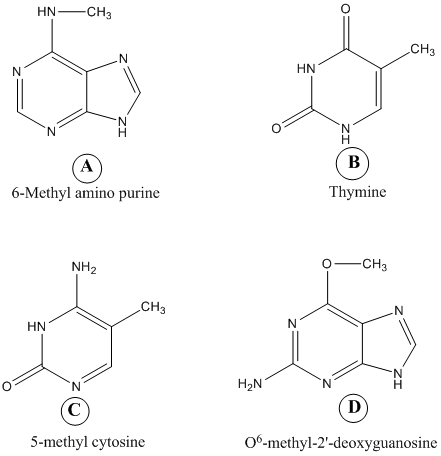 The following are the points related to any one or two of the above given structures. The points along with the name of the structure labeled from A to D are as follows:
The following are the points related to any one or two of the above given structures. The points along with the name of the structure labeled from A to D are as follows:
(a)In deoxyribonucleic acid (DNA) mismatch repair mechanism the errors in newly synthesized DNA strand is identified and corrected. To distinguish parent strand from daughter strand it adopts hemimethylation. In hemimethylation, the parent strand exists in methylated form while daughter strand is left unmethylated. Hence, 6- Methyl amino purine helps in identifying parental DNA strand in the Mut H, L, S mismatch correction system.
Hence, the characteristic (a) is followed by structure a.(b)Ultraviolet (UV) light induces pyrimidine dimers formation in the DNA strand. The different types of dimers include cyclobutane pyrimidine dimer, 6,4 pyrimidine-pyrimidones. Thymine is a pyrimidine which is most likely to be involved in cyclobutane dimer formation after ultraviolet irradiation of DNa.Hence, the characteristic (b) is followed by structure b.(c)5-methyl cytosine is a methylated base found immediately to the 5' side of dGMP (deoxy guaanosine monophosphate) residues in eukaryotic DNa.Hence, the characteristic (c) is followed by structure c.(d)Nucleotide excision repair mechanism removes DNA damaged by UV radiations. Some chemical agents when treated with DNA they alkylate bases.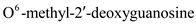 is created by treating DNA with alkylating agents that transfer methyl groups and repaired by an enzyme that functions only once in its lifetime.
is created by treating DNA with alkylating agents that transfer methyl groups and repaired by an enzyme that functions only once in its lifetime.
Hence, the characteristic (d) is followed by structure D.
(e)6- Methyl amino purine and 5-methyl cytosine are created by S-adenosyl-L-methionine (S-AdoMet or SAM) dependent methylation of nucleotide residue in DNA. S-AdoMet is a co-substrate involved in methyl group transfers
Hence, the characteristic (e) is followed by structure A and c.(f)5-methyl cytosine is a substrate for deamination at the DNA level which would lead to a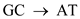 transition. Deamination is a process to repair DNA. 5-methyl cytosine is formed by treatment with SAM.
transition. Deamination is a process to repair DNA. 5-methyl cytosine is formed by treatment with SAM.
Hence, the characteristic (f) is followed by structure c.
 The following are the points related to any one or two of the above given structures. The points along with the name of the structure labeled from A to D are as follows:
The following are the points related to any one or two of the above given structures. The points along with the name of the structure labeled from A to D are as follows:(a)In deoxyribonucleic acid (DNA) mismatch repair mechanism the errors in newly synthesized DNA strand is identified and corrected. To distinguish parent strand from daughter strand it adopts hemimethylation. In hemimethylation, the parent strand exists in methylated form while daughter strand is left unmethylated. Hence, 6- Methyl amino purine helps in identifying parental DNA strand in the Mut H, L, S mismatch correction system.
Hence, the characteristic (a) is followed by structure a.(b)Ultraviolet (UV) light induces pyrimidine dimers formation in the DNA strand. The different types of dimers include cyclobutane pyrimidine dimer, 6,4 pyrimidine-pyrimidones. Thymine is a pyrimidine which is most likely to be involved in cyclobutane dimer formation after ultraviolet irradiation of DNa.Hence, the characteristic (b) is followed by structure b.(c)5-methyl cytosine is a methylated base found immediately to the 5' side of dGMP (deoxy guaanosine monophosphate) residues in eukaryotic DNa.Hence, the characteristic (c) is followed by structure c.(d)Nucleotide excision repair mechanism removes DNA damaged by UV radiations. Some chemical agents when treated with DNA they alkylate bases.
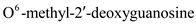 is created by treating DNA with alkylating agents that transfer methyl groups and repaired by an enzyme that functions only once in its lifetime.
is created by treating DNA with alkylating agents that transfer methyl groups and repaired by an enzyme that functions only once in its lifetime.Hence, the characteristic (d) is followed by structure D.
(e)6- Methyl amino purine and 5-methyl cytosine are created by S-adenosyl-L-methionine (S-AdoMet or SAM) dependent methylation of nucleotide residue in DNA. S-AdoMet is a co-substrate involved in methyl group transfers
Hence, the characteristic (e) is followed by structure A and c.(f)5-methyl cytosine is a substrate for deamination at the DNA level which would lead to a
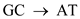 transition. Deamination is a process to repair DNA. 5-methyl cytosine is formed by treatment with SAM.
transition. Deamination is a process to repair DNA. 5-methyl cytosine is formed by treatment with SAM.Hence, the characteristic (f) is followed by structure c.
4
Homologous recombination in E. coli forms heteroduplex regions of DNA containing mismatched bases. Why are these mismatches not eliminated by the mismatch repair system

Unlock Deck
Unlock for access to all 11 flashcards in this deck.
Unlock Deck
k this deck
5
Deficiencies in the activity of either dUTPase or DNA ligase stimulate recombination. Why

Unlock Deck
Unlock for access to all 11 flashcards in this deck.
Unlock Deck
k this deck
6
Suppose that you wanted to study retroviral integration mechanisms by determining the nucleotide sequence at the integration site-several dozen nucleotides on each side of the viral-cellular DNA junction. Describe how to isolate DNA containing a junction site in amounts sufficient for sequence analysis.

Unlock Deck
Unlock for access to all 11 flashcards in this deck.
Unlock Deck
k this deck
7
Analysis of p53 gene mutations in human tumors shows that a large proportion of these mutations involve GC AT transitions originating at sites of DNA methylation. Propose a model to explain preferential mutagenesis of this type at these sites.

Unlock Deck
Unlock for access to all 11 flashcards in this deck.
Unlock Deck
k this deck
8
A paper in the Journal of Bacteriology reported that a mutT mutant strain of E. coli displayed a mutator phenotype when the bacteria were cultured anaerobically. Is this a surprising result Briefly explain your answer. Present one or two possible explanations for this observation.

Unlock Deck
Unlock for access to all 11 flashcards in this deck.
Unlock Deck
k this deck
9
It is thought that the signal for eukaryotic mismatch repair, for identifying the strand to be repaired, could be nicks in the DNA to be repaired. If so, would you expect the nicked strand or the intact strand to be the strand to be repaired Briefly explain your answer.

Unlock Deck
Unlock for access to all 11 flashcards in this deck.
Unlock Deck
k this deck
10
Identify and briefly describe three of the processes by which deamination of DNA-cytosine residues by AID could lead to mutagenesis.

Unlock Deck
Unlock for access to all 11 flashcards in this deck.
Unlock Deck
k this deck
11
In what ways can insertion of a transposon affect the expression of genes in the neighborhood of the insertion site

Unlock Deck
Unlock for access to all 11 flashcards in this deck.
Unlock Deck
k this deck


