Deck 26: Information Readout: Transcription and Post-Transcriptional Processing
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/14
Play
Full screen (f)
Deck 26: Information Readout: Transcription and Post-Transcriptional Processing
1
Outline an experimental approach to determining the average chain growth rate for transcription in vivo. Chain growth rate is the number of nucleotides polymerized per minute per RNA chain.
The transcription is the process of copying the DNA (deoxyribonucleic acid) segment into RNA (ribonucleic acid). The chain growth is the number of nucleotides added to RNA chain per minute. The average rate of chain growth can be determined experimentally in vivo as follows:
• The RNA (ribonucleic acid) precursor should be labeled as radioactive and isolated. This RNA should then be treated with mild alkali. This treatment will give one nucleoside at the 3' terminal. To calculate the chain growth rate divide the total amount of labeled nucleotide to the amount of nucleosides formed.
• The RNA (ribonucleic acid) precursor should be labeled as radioactive and isolated. This RNA should then be treated with mild alkali. This treatment will give one nucleoside at the 3' terminal. To calculate the chain growth rate divide the total amount of labeled nucleotide to the amount of nucleosides formed.
2
Outline an experimental approach to demonstrate the average RNA chain growth rate during transcription of a cloned gene in vitro.
The chain growth is the number of nucleotides added to RNA (ribonucleic acid) chain per minute. The chain growth rate of transcription in vitro can be calculated as follows:
• The conditions are set up such that cloned gene is allowed to synthesize. The transcription is initiated by adding three ribonucleotides. When the time is started to note down, at that time the fourth nucleotide is added. The reaction is stopped after a certain limit of time. The template is added which would help to measure the amount of transcribing genes in the experiment. This could be determined by gel electrophoresis.
• The conditions are set up such that cloned gene is allowed to synthesize. The transcription is initiated by adding three ribonucleotides. When the time is started to note down, at that time the fourth nucleotide is added. The reaction is stopped after a certain limit of time. The template is added which would help to measure the amount of transcribing genes in the experiment. This could be determined by gel electrophoresis.
3
Measurements of RNA chain growth rates are often led astray by the phenomenon of pausing, in which an RNA polymerase molecule stops transcription when it reaches certain sites, for intervals that may be as long as several minutes. How might pausing be detected
The pausing can be detected by gel electrophoresis. In the gel electrophoresis, the paused species of RNA (ribonucleic acid) will accumulate and appear as heavy RNA (ribonucleic acid) particles, which are not observed generally.
4
Suppose you want to study the transcription in vitro of one particular gene in a DNA molecule that contains several genes and promoters. Without adding specific regulatory proteins, how might you stimulate transcription from the gene of interest relative to the transcription of the other genes on your DNA template To make all of the complexes identical, you would like to arrest all transcriptional events at the same position on the DNA template before isolating the complex. How might you do this

Unlock Deck
Unlock for access to all 14 flashcards in this deck.
Unlock Deck
k this deck
5
The tac promoter, an artificial promoter made from a chemically synthesized oligonucleotide, has been introduced into a plasmid. It is a hybrid of the lac and trp promoters, containing the 35 region of one and the 10 region of the other. This promoter directs transcription initiation more efficiently than either the trp or lac promoters. Why

Unlock Deck
Unlock for access to all 14 flashcards in this deck.
Unlock Deck
k this deck
6
Would you expect actinomycin D to be a competitive inhibitor of RNA polymerase What about cordycepin Briefly explain your answers.

Unlock Deck
Unlock for access to all 14 flashcards in this deck.
Unlock Deck
k this deck
7
A restriction fragment was subjected to Maxam-Gilbert sequencing, with results as shown in the first four lanes of the radioautogram. S 1 nuclease mapping was carried out, for a gene whose transcription initiated within this sequence (see Tools of Biochemistry 27B). The transcript protected a fragment whose length was as shown in the fifth lane. 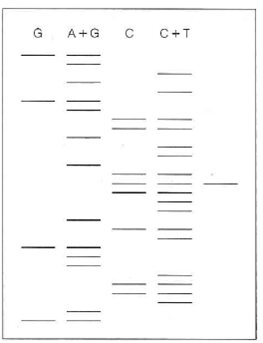
(a) Give the nucleotide sequence of both DNA strands and the first several RNA nucleotides. Identify all 3 and 5 ends.
(b) On your structure, show the approximate location of the Pribnow box (the -10 region).
(c) Assuming that the restriction fragment was created with a type II restriction endonuclease that recognizes a 6-base-pair site, show the structure of that site, and indicate where cleavage occurs.

(a) Give the nucleotide sequence of both DNA strands and the first several RNA nucleotides. Identify all 3 and 5 ends.
(b) On your structure, show the approximate location of the Pribnow box (the -10 region).
(c) Assuming that the restriction fragment was created with a type II restriction endonuclease that recognizes a 6-base-pair site, show the structure of that site, and indicate where cleavage occurs.

Unlock Deck
Unlock for access to all 14 flashcards in this deck.
Unlock Deck
k this deck
8
Explain the basis for the following statement: Transcription of two genes on a plasmid can occur without the concomitant action of a topoisomerase, but only if those two genes are oriented in opposite directions.

Unlock Deck
Unlock for access to all 14 flashcards in this deck.
Unlock Deck
k this deck
9
Some years ago, it was suggested that the function of the poly(A) tail on a eukaryotic message may be to "ticket" the message. That is, each time the message is used, one or more residues is removed, and the message is degraded after the tail is shortened below a critical length. Suggest an experiment to test this hypothesis.

Unlock Deck
Unlock for access to all 14 flashcards in this deck.
Unlock Deck
k this deck
10
It has been proposed that nucleosomes must be removed in order for transcription to proceed through chromatin. Suggest an experiment that might test this hypothesis.

Unlock Deck
Unlock for access to all 14 flashcards in this deck.
Unlock Deck
k this deck
11
Shown below is an R loop prepared for electron microscopy by
annealing a purified eukaryotic messenger RNA with DNA from a genomic clone containing the full-length gene corresponding to the mRNA.
(a) How many exons does the gene contain How many introns
(b) Where in this structure would you expect to find a 5 ,5 -internucleotide bond Where would you expect to find a polyadenylic acid sequence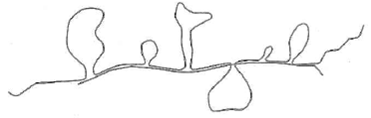
annealing a purified eukaryotic messenger RNA with DNA from a genomic clone containing the full-length gene corresponding to the mRNA.
(a) How many exons does the gene contain How many introns
(b) Where in this structure would you expect to find a 5 ,5 -internucleotide bond Where would you expect to find a polyadenylic acid sequence


Unlock Deck
Unlock for access to all 14 flashcards in this deck.
Unlock Deck
k this deck
12
Introns in eukaryotic protein-coding genes are rarely shorter than 65 nucleotides in length. What might be a rationale for this limitation

Unlock Deck
Unlock for access to all 14 flashcards in this deck.
Unlock Deck
k this deck
13
Heparin is a polyanionic polysaccharide that blocks elongation by RNA polymerase. But heparin inhibits only when added before the onset of transcription and not if added after transcription begins. Explain this difference.

Unlock Deck
Unlock for access to all 14 flashcards in this deck.
Unlock Deck
k this deck
14
Estimate the time needed for the E. coli RNA polymerase at 37 °C to transcribe the entire gene for a 50-kilodalton protein. What assumption or assumptions must be made for this estimate to be accurate

Unlock Deck
Unlock for access to all 14 flashcards in this deck.
Unlock Deck
k this deck


