Deck 12: Chromatin Structure and Its Effectson Transcription
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/29
Play
Full screen (f)
Deck 12: Chromatin Structure and Its Effectson Transcription
1
If the globin locus did have the same DNase-hypersensitive sites in J6 cells as in HEL cells, approximately what size fragments would have been detected in Figure 13.20d Which hypersensitive sites would not be detected
Reference Figure 13.20d: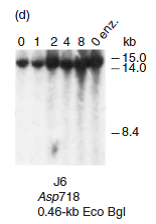
Reference Figure 13.20d:

Yaniv and his colleagues secluded nuclei from SV40 virus diseased monkey cells at 44, 24, 34 h after the injection and treated them having DNase I. Then they reduced the minichromosomes which are treated having EcoRI and examined the DNA products through searching, electrophoresis and Southern staining having SV40 DNA which is radioactive.
The chromatin's region which are sensitive to cleavage through deoxyribonuclease (DNase) enzymes are known as DNase hypersensitive sites. If the globin locus in J6 cells had the similar pattern of DNase hypersensitivity as it does in HEL cells, then the results in panel (d) would be identical to those in panel (a). This is because the same restriction endonuclease was used to recut the DNA after DNase I treatment, and the probes (0.46 Eco Bgl and 1.4-kb Bam would hybridize to the same fragments created by DNase cleavage at sites 3a, 3b, and 4.
The chromatin's region which are sensitive to cleavage through deoxyribonuclease (DNase) enzymes are known as DNase hypersensitive sites. If the globin locus in J6 cells had the similar pattern of DNase hypersensitivity as it does in HEL cells, then the results in panel (d) would be identical to those in panel (a). This is because the same restriction endonuclease was used to recut the DNA after DNase I treatment, and the probes (0.46 Eco Bgl and 1.4-kb Bam would hybridize to the same fragments created by DNase cleavage at sites 3a, 3b, and 4.
2
Diagram a nucleosome as follows:
(a) On a drawing of the histones without the DNA, show the rough positions of all the histones. (b) On a separate drawing, show the path of DNA around the histones.
(a) On a drawing of the histones without the DNA, show the rough positions of all the histones. (b) On a separate drawing, show the path of DNA around the histones.
Histones are the proteins that are located in the nucleus of eukaryotic cells. There are five different kinds of histones - H1, H2A, H2B, H3 and H4. The four histone proteins out of these five - H2A, H2B, H3 and H4 together act as spools around which the DNA is wound. The DNA wound on to the core protein form the structure known as nucleosome.
Based on the x-ray diffraction studies, scientist Baldwin deduced the structure of nucleosome. The core proteins (four histone proteins) form a ball and the DNA is wrapped on the outside. The location of DNA outside the spool minimizes the bending of DNA. The two H2A-H2B dimers are attached to the central core of (H3-H4)2 tetramer forming a ball of histone octane structure. The histone protein H1 is located on the outside
Diagrammatic representation of nucleosome - (a) histones without DNA wrapped around and b. Histones with DNA -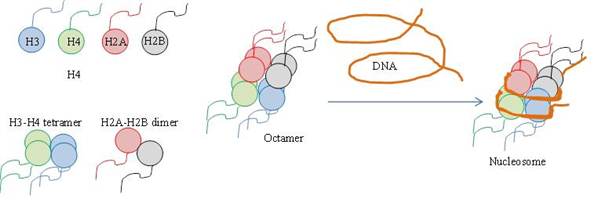 Thus, nucleosome makes the long DNA compact and can fit it into the nucleus of the cell.
Thus, nucleosome makes the long DNA compact and can fit it into the nucleus of the cell.
Based on the x-ray diffraction studies, scientist Baldwin deduced the structure of nucleosome. The core proteins (four histone proteins) form a ball and the DNA is wrapped on the outside. The location of DNA outside the spool minimizes the bending of DNA. The two H2A-H2B dimers are attached to the central core of (H3-H4)2 tetramer forming a ball of histone octane structure. The histone protein H1 is located on the outside
Diagrammatic representation of nucleosome - (a) histones without DNA wrapped around and b. Histones with DNA -
 Thus, nucleosome makes the long DNA compact and can fit it into the nucleus of the cell.
Thus, nucleosome makes the long DNA compact and can fit it into the nucleus of the cell. 3
Explain why brief digestion of eukaryotic chromatin with micrococcal nuclease gives DNA fragments about 200 bp long, but longer digestion yields 146-bp fragments.
Brief digestion of eukaryotic chromatin with micrococcal nuclease yields DNA fragments about 200 bp long because this mild digestion cuts only the most nuclease-sensitive sites, which lie in the inter-nucleosomal regions, and are spaced about 200 bp apart. Longer digestion yields DNA fragments that are about 147 bp long because this more stringent treatment digests all the DNA that is not protected by the nucleosomal proteins, and that protected region spans about 147 base pairs.
4
Cite electron microscopic evidence for a six- to seven fold condensation of DNA in nucleosomes.

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
5
The amino acid sequences of the core histones are highly conserved between plants and animals. Present a hypothesis to explain this finding.

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
6
Cite electron microscopic evidence for formation of a condensed fiber (30-nm fiber) at high ionic strength

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
7
Type A histone acetyltransferases (HAT A's) contain a bromodomain and HAT B's do not. What do you predict would occur if HAT A's were missing this bromodomain What if HAT B's possessed this bromodomain If the bromodomains were reversed so that all HAT B's gained bromodomains and all HAT A's lost them, would HAT A's take over the role of HAT B's and vice versa Why or why not How would you answer this question experimentally

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
8
Diagram the solenoid model of the 30-nm chromatin fiber

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
9
Diagram the structure of a tetranucleosome revealed by x-ray crystallography. What structure for the 30-nm fiber does this tetranucleosome structure suggest

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
10
How can single-molecule force spectrospcopy shed light on the structure of the 30-nm chromatin fiber What conclusions does it suggest

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
11
Draw a model to explain the next order of chromatin folding after the 30-nm fiber. Cite biochemical and microscopic evidence to support the mode

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
12
Describe and give the results of an experiment that shows the competing effects of histone H1 and the activator Gal4-VP16 on transcription of the adenovirus E4 gene in reconstituted chromatin.

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
13
Present two models for antirepression by transcription activators, one in which the gene's control region is not blocked by a nucleosome, the other in which it is.

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
14
Describe and give the results of an experiment that shows that the nucleosome-free zone in active SV40 chromatin lies at the viral late gene control region

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
15
Describe and give the results of an experiment that shows that the zone of DNase hypersensitivity in SV40 chromatin lies at the viral late gene control region

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
16
Diagram and describe a general technique for detecting a DNase-hypersensitive DNA region

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
17
Describe and give the results of an activity gel assay that shows the existence of a histone acetyltransferase (HAT) activity.

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
18
Present a model for the involvement of a corepressor and histone deacetylase in transcription repression.

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
19
Describe and give the results of an epitope-tagging experiment that shows interaction among the following three proteins, the repressor Mad1, the corepressor SIN3A, and the histone deacetylase HDAC2

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
20
Present a model for activation and repression by the same protein, depending upon the presence or absence of that protein's ligand.

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
21
Present models for uncatalyzed nucleosomal DNA exposure and for catalyzed nucleosomal remodeling. Present evidence for the catalyzed model

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
22
Describe how you could use a chromatin immunoprecipitation procedure to detect the proteins associated with a particular gene at various points in the cell cycle.

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
23
Describe and give the results of an experiment using chromatin immunoprecipitation to discover the timing of acetylation and phosphorylation of particular sites on core histones in a nucleosome at the IFN- promoter.

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
24
Describe and give the results of an experiment to measure recruitment of SWI/SNF and TFIID to the IFN- promoter with wild-type and mutant histones

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
25
Present a model depicting the establishment and decoding of a histone code at the IFN- promoter

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
26
Present a model to explain why lysine 16 in histone H4 is thought to be critical for silencing. What evidence supports this hypothesis

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
27
Present a model depicting the spread of chromatin repression via histone methylation

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
28
Present a model of the interactions among the modifications of lysines 9 and 14, and serine 10 in the N-terminal tail of histone H3. Show both positive and negative interactions.

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
29
Present evidence that FACT causes a loss of histone H2A-H2B dimers from nucleosomes, and that this activity depends on the C-terminus of the Spt16 subunit of FACT

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck


