Deck 1: DNA: The Genetic Material
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/43
Play
Full screen (f)
Deck 1: DNA: The Genetic Material
1
Griffith's experiment injecting a mixture of dead and live bacteria into mice demonstrated that (choose the correct answer) a. DNA is double-stranded.
B) mRNA of eukaryotes differs from mRNA of prokaryotes.
C) a factor was capable of transforming one bacterial cell type to another.
D) bacteria can recover from heat treatment if live helper cells are present.
B) mRNA of eukaryotes differs from mRNA of prokaryotes.
C) a factor was capable of transforming one bacterial cell type to another.
D) bacteria can recover from heat treatment if live helper cells are present.
In 1928, a British medical officer by the name of Frederick Griffith conducted an experiment using live and dead bacteria. He used two strains of Streptococcus pneumonia , the R strain and the S strain. The R strain is rough and nonvirulent, while the S strain is smooth and virulent. In order to further facilitate his experiment, Griffith used two different types of the bacteria, those with type II coats and those with type III coats.
Thus, Griffith used Type IIR and Type IIIS bacteria. Since bacteria can mutate, it is possible for R bacteria to mutate into S and vice versa; however, there is no mutation between types. Type IIR bacteria can mutate into Type IIS bacteria but not into any Type III bacteria. Similarly, Type IIIR bacteria can mutate into type IIIS bacteria but not into any type II bacteria.
Griffith injected combinations of the living and dead bacteria in to mice. When Griffith injected Type IIR bacteria in to the mice, the mice survived with no bacteria present in their bloodstream. When Griffith injected Type IIIS bacteria in to mice, the mice died with virulent bacteria found in their bloodstream. When Griffith injected heat-killed Type IIIS bacteria in to mice, the mice survived with no bacteria present in their bloodstream. Finally, when Griffith injected Type IIR bacteria and heat killed Type IIIS bacteria into mice, the mice died with Type IIIS bacteria found in their bloodstream.
Since, in the last experiment, Griffith knew that Type IIR bacteria cannot mutate in to Type IIIS bacteria, he concluded that some of the Type IIR bacteria had been transformed into Type IIIS bacteria via interaction between the dead and live cells.
Choice (a) is incorrect because the experimental results do not indicate whether or not the genetic material is deoxyribonucleic acid (DNA) or protein. So, it does not indicate that the DNA is double stranded. In fact, Griffith believed that protein acted as the genetic material, not DNA.
Thus, choice (a) is incorrect.
Choice (b) is incorrect because Griffith's experiment is not at all about messenger ribonucleic acid (mRNA). Griffith's experiment is about how genetic information is passed between bacterial strains, which are prokaryotes.
Thus, choice (b) is incorrect.
Choice (d) is incorrect because there is no such thing as helper cells for bacteria. Additionally, bacteria cannot recover from heat treatment but they can transfer their DNA to other strains.
Thus, choice (d) is incorrect.
Choice (c) is correct because Griffith did show that there is a factor that transforms one bacterial cell type into another.
Hence, the correct answer is:
(c) a factor was capable of transforming one bacterial cell type to another.
Thus, Griffith used Type IIR and Type IIIS bacteria. Since bacteria can mutate, it is possible for R bacteria to mutate into S and vice versa; however, there is no mutation between types. Type IIR bacteria can mutate into Type IIS bacteria but not into any Type III bacteria. Similarly, Type IIIR bacteria can mutate into type IIIS bacteria but not into any type II bacteria.
Griffith injected combinations of the living and dead bacteria in to mice. When Griffith injected Type IIR bacteria in to the mice, the mice survived with no bacteria present in their bloodstream. When Griffith injected Type IIIS bacteria in to mice, the mice died with virulent bacteria found in their bloodstream. When Griffith injected heat-killed Type IIIS bacteria in to mice, the mice survived with no bacteria present in their bloodstream. Finally, when Griffith injected Type IIR bacteria and heat killed Type IIIS bacteria into mice, the mice died with Type IIIS bacteria found in their bloodstream.
Since, in the last experiment, Griffith knew that Type IIR bacteria cannot mutate in to Type IIIS bacteria, he concluded that some of the Type IIR bacteria had been transformed into Type IIIS bacteria via interaction between the dead and live cells.
Choice (a) is incorrect because the experimental results do not indicate whether or not the genetic material is deoxyribonucleic acid (DNA) or protein. So, it does not indicate that the DNA is double stranded. In fact, Griffith believed that protein acted as the genetic material, not DNA.
Thus, choice (a) is incorrect.
Choice (b) is incorrect because Griffith's experiment is not at all about messenger ribonucleic acid (mRNA). Griffith's experiment is about how genetic information is passed between bacterial strains, which are prokaryotes.
Thus, choice (b) is incorrect.
Choice (d) is incorrect because there is no such thing as helper cells for bacteria. Additionally, bacteria cannot recover from heat treatment but they can transfer their DNA to other strains.
Thus, choice (d) is incorrect.
Choice (c) is correct because Griffith did show that there is a factor that transforms one bacterial cell type into another.
Hence, the correct answer is:
(c) a factor was capable of transforming one bacterial cell type to another.
2
In the 1920s, while working with Streptococcus pneumoniae (the agent that causes pneumonia), Griffith injected mice with different types of bacteria. For each of the following bacteria types injected, indicate whether the mice lived or died:
a. type IIR
b. type IIIS
c. heat-killed IIIS
d. type IIR + heat-killed IIIS
a. type IIR
b. type IIIS
c. heat-killed IIIS
d. type IIR + heat-killed IIIS
In 1928, a British medical officer by the name of Frederick Griffith conducted an experiment using live and dead bacteria. He used two strains of Streptococcus pneumonia , the R strain and the S strain. The R strain is rough and nonvirulent, while the S strain is smooth and virulent. In order to further facilitate his experiment, Griffith used two different types of the bacteria, those with type II coats and those with type III coats.
Thus, Griffith used Type IIR and Type IIIS bacteria. Since bacteria can mutate, it is possible for R bacteria to mutate into S and vice versa; however, there is not mutation between types. Type IIR bacteria can mutate into Type IIS bacteria but not into any Type III bacteria. Similarly, Type IIIR bacteria can mutate into type IIIS bacteria but not into any type II bacteria.
a) When Griffith injected Type IIR bacteria in to the mice, the mice survived with no bacteria present in their bloodstream. The mice survived because they were infected with the nonvirulent strain and there was no way for it to become virulent.
b) When Griffith injected Type IIIS bacteria in to mice, the mice died with virulent bacteria found in their bloodstream. The mice died because they were infected with the live virulent strain.
c) When Griffith injected heat-killed Type IIIS bacteria in to mice, the mice survived with no bacteria present in their bloodstream. The mice survived because they were infected with a dead strain, so it didn't matter if it was previously virulent or not.
d) Finally, when Griffith injected Type IIR bacteria and heat killed Type IIIS bacteria into mice, the mice died with Type IIIS bacteria found in their bloodstream. The mice died because the nonvirulent strain was transformed into the virulent strain through its interaction with the dead virulent strain.
Thus, Griffith used Type IIR and Type IIIS bacteria. Since bacteria can mutate, it is possible for R bacteria to mutate into S and vice versa; however, there is not mutation between types. Type IIR bacteria can mutate into Type IIS bacteria but not into any Type III bacteria. Similarly, Type IIIR bacteria can mutate into type IIIS bacteria but not into any type II bacteria.
a) When Griffith injected Type IIR bacteria in to the mice, the mice survived with no bacteria present in their bloodstream. The mice survived because they were infected with the nonvirulent strain and there was no way for it to become virulent.
b) When Griffith injected Type IIIS bacteria in to mice, the mice died with virulent bacteria found in their bloodstream. The mice died because they were infected with the live virulent strain.
c) When Griffith injected heat-killed Type IIIS bacteria in to mice, the mice survived with no bacteria present in their bloodstream. The mice survived because they were infected with a dead strain, so it didn't matter if it was previously virulent or not.
d) Finally, when Griffith injected Type IIR bacteria and heat killed Type IIIS bacteria into mice, the mice died with Type IIIS bacteria found in their bloodstream. The mice died because the nonvirulent strain was transformed into the virulent strain through its interaction with the dead virulent strain.
3
In the key transformation experiment performed by Griffith, mice were injected with living IIR bacteria mixed with heat-killed IIIS bacteria.
a. What type of bacteria were recovered?
b. What result would you expect if living IIIR bacteria had been mixed with heat-killed IIS bacteria?
c. Explain why, for Griffith to interpret his results as evidence of transformation, it was necessary for him to mix living IIR bacteria with dead IIIS bacteria and not with dead IIS bacteria.
a. What type of bacteria were recovered?
b. What result would you expect if living IIIR bacteria had been mixed with heat-killed IIS bacteria?
c. Explain why, for Griffith to interpret his results as evidence of transformation, it was necessary for him to mix living IIR bacteria with dead IIIS bacteria and not with dead IIS bacteria.
In 1928, a British medical officer by the name of Frederick Griffith conducted an experiment using live and dead bacteria. He used two strains of Streptococcus pneumonia , the R strain and the S strain. The R strain is rough and nonvirulent, while the S strain is smooth and virulent. In order to further facilitate his experiment, Griffith used two different types of the bacteria, those with type II coats and those with type III coats.
Thus, Griffith used Type IIR and Type IIIS bacteria. Since bacteria can mutate, it is possible for R bacteria to mutate into S and vice versa; however, there is no mutation between types. Type IIR bacteria can mutate into Type IIS bacteria but not into any Type III bacteria. Similarly, Type IIIR bacteria can mutate into type IIIS bacteria but not into any type II bacteria.
a) When Griffith injected Type IIR bacteria and heat killed Type IIIS bacteria into mice, the mice died with Type IIIS bacteria found in their bloodstream. The mice died because the nonvirulent strain was transformed into the virulent strain through its interaction with the dead virulent strain.
b) If living IIIR bacteria have been mixed with heat killed IIS bacteria, the expectation would be that some of the IIIR bacteria would transform into IIS bacteria by picking up the genetic material of the dead IIS bacteria. If the combination were injected into a mouse, the mouse would die. Though it could fight off the IIIR bacteria, the mouse would not be able to fight off the IIS bacteria. The IIS bacteria would be found in the bloodstream of the dead mouse.
c) In order for Griffith to interpret his results as transformation, he had to mix IIR bacteria with IIIS bacteria.
If Griffith had mixed IIR bacteria with IIS bacteria, he could not have unequivocally concluded that transformation had occurred because it would have also been possible that the IIR strain could have mutated in to the IIS strain.
Thus, Griffith used Type IIR and Type IIIS bacteria. Since bacteria can mutate, it is possible for R bacteria to mutate into S and vice versa; however, there is no mutation between types. Type IIR bacteria can mutate into Type IIS bacteria but not into any Type III bacteria. Similarly, Type IIIR bacteria can mutate into type IIIS bacteria but not into any type II bacteria.
a) When Griffith injected Type IIR bacteria and heat killed Type IIIS bacteria into mice, the mice died with Type IIIS bacteria found in their bloodstream. The mice died because the nonvirulent strain was transformed into the virulent strain through its interaction with the dead virulent strain.
b) If living IIIR bacteria have been mixed with heat killed IIS bacteria, the expectation would be that some of the IIIR bacteria would transform into IIS bacteria by picking up the genetic material of the dead IIS bacteria. If the combination were injected into a mouse, the mouse would die. Though it could fight off the IIIR bacteria, the mouse would not be able to fight off the IIS bacteria. The IIS bacteria would be found in the bloodstream of the dead mouse.
c) In order for Griffith to interpret his results as transformation, he had to mix IIR bacteria with IIIS bacteria.
If Griffith had mixed IIR bacteria with IIS bacteria, he could not have unequivocally concluded that transformation had occurred because it would have also been possible that the IIR strain could have mutated in to the IIS strain.
4
Several years after Griffith described the transforming principle, Avery, MacLeod, and McCarty investigated the same phenomenon.
a. List the steps they used to show that DNA from dead S. pneumoniae cells was responsible for the change from a nonvirulent to a virulent state.
b. What was the role of enzymes in these experiments?
c. Did their work confirm or disconfirm Griffith's work, and how?
a. List the steps they used to show that DNA from dead S. pneumoniae cells was responsible for the change from a nonvirulent to a virulent state.
b. What was the role of enzymes in these experiments?
c. Did their work confirm or disconfirm Griffith's work, and how?

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
5
Hershey and Chase showed that when phages were labeled witH32 P and 35 S, the 35 S remained outside the cell and could be removed without affecting the course of infection, whereas the 32 P entered the cell and could be recovered in progeny phages.
a. What distribution of isotopes would you expect to see if parental phages were labeled with isotopes of
i. C?
ii. N?
iii. H?
b. Based on your answer, explain why Hershey and Chase used isotopes of phosphorus and sulfur in their experiments.
a. What distribution of isotopes would you expect to see if parental phages were labeled with isotopes of
i. C?
ii. N?
iii. H?
b. Based on your answer, explain why Hershey and Chase used isotopes of phosphorus and sulfur in their experiments.

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
6
Suppose you identify a previously unknown multicellular organism.
a. What composition do you expect its genome to have?
b. How would your answer change if it were a unicellular organism?
c. How would your answer change if it were a bacteriophage or virus?
d. Do your answers offer any insights into the origins of cellular organisms?
a. What composition do you expect its genome to have?
b. How would your answer change if it were a unicellular organism?
c. How would your answer change if it were a bacteriophage or virus?
d. Do your answers offer any insights into the origins of cellular organisms?

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
7
How could you use radioactively labeled molecules to determine if the genome of a newly identified bacteriophage that infects E. coli is RNA or DNA? How might you determine if it is composed of single-stranded or double stranded nucleic acid?

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
8
The X-ray diffraction data obtained by Rosalind Franklin suggested that (choose the correct answer)a. DNA is a helix with a pattern that repeats every 3.4 nm.
b. purines are hydrogen bonded to pyrimidines.
c. DNA is a left-handed helix.
d. DNA is organized into nucleosomes.
b. purines are hydrogen bonded to pyrimidines.
c. DNA is a left-handed helix.
d. DNA is organized into nucleosomes.

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
9
What evidence do we have that, in the helical form of the DNA molecule, the base pairs are composed of one purine and one pyrimidine?

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
10
What exactly is a deoxyribonucleotide made up of, and how many different deoxyribonucleotides are there in DNA? Describe the structure of DNA, and describe the onding mechanism of the molecule (i.e., the kind of bonds on the sides of the "ladder" and the kind of bonds holding the two complementary strands together). Base pairing in DNA consists of purine-pyrimidine pairs, so why is it not possible for A-C and G-T pairs to form?

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
11
What is the base sequence, given to , of the DNA strand that would be complementary to the following single-stranded DNA molecules?
a. 5'-AGTTACCTGATGGTA- 3'
b. 5'-TTCTCAAGAATTCCA- 3'
a. 5'-AGTTACCTGATGGTA- 3'
b. 5'-TTCTCAAGAATTCCA- 3'

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
12
The phosphodiester bonds that lie exactly in the iddle of an 8-bp long segment of double-stranded DNA are broken to create two 4-bp long molecules. Phosphodiester bonds between the resulting two doublestranded molecules are then reformed, but without regard to their initial order. For each of the following sequences (the sequence given is that of just one strand), list all possible double-stranded sequences that can be formed.
a. 5'-TTAACCGG-3' (on this strand, the phosphodiester bond between A and C is broken)b. 5'-TTCCAAGG-3'(on this strand, the phosphodiester bond between C and A is broken)c. 5'-AGCTAGCT-3' (on this strand, the phosphodiester bond between T and A is broken)d. 5'-AGCTTCGA-3' (on this strand, the phosphodiester bond between the two Ts is broken)
a. 5'-TTAACCGG-3' (on this strand, the phosphodiester bond between A and C is broken)b. 5'-TTCCAAGG-3'(on this strand, the phosphodiester bond between C and A is broken)c. 5'-AGCTAGCT-3' (on this strand, the phosphodiester bond between T and A is broken)d. 5'-AGCTTCGA-3' (on this strand, the phosphodiester bond between the two Ts is broken)

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
13
Describe the bonding properties of G-C and T-A. Which base pair would be harder to break apart? Why?

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
14
The double-helix model of DNA, as suggested by Watson and Crick, was based on DNA data gathered by other researchers. The facts fell into the following two general categories:
a. chemical composition
b. physical structure
Give two examples of each.
a. chemical composition
b. physical structure
Give two examples of each.

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
15
For double-stranded DNA, which of the following base ratios always equals 1?
a. ( A+T )/( G+C )b. ( A+G )/( C+T )c. C / G
d. ( G+T )/( A+C )e. A / G
a. ( A+T )/( G+C )b. ( A+G )/( C+T )c. C / G
d. ( G+T )/( A+C )e. A / G

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
16
Suppose the ratio of ( A + T ) to ( G + C ) in a particular DNA is 1.0. Does this ratio indicate that the DNA is probably composed of two complementary strands of DNA, or a single strand of DNA, or is more information necessary?

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
17
The percentage of cytosine in a double-stranded DNA is 17. What is the percentage of adenine in that DNA?

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
18
A double-stranded DNA polynucleotide contains 80 thymidylic acid and 110 deoxyguanylic acid residues. What is the total nucleotide number in this DNA fragment?

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
19
Analysis of DNA from a bacterial virus indicates hat it contains 33% A , 26% T , 18% G , and 23% C. Interpret these data.

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
20
The following are melting temperatures for different double-stranded DNA molecules:
a. 73°C
b. 69°C
c. 84°C
d. 78°C
e. 82°C
Arrange these molecules from lower to higher content of G-C pairs.
a. 73°C
b. 69°C
c. 84°C
d. 78°C
e. 82°C
Arrange these molecules from lower to higher content of G-C pairs.

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
21
coli bacteriophage X174 and parvovirus B19 (the causative agent of Fifth disease- infectious redness -in humans) each have a single-stranded DNA genome.
a. What base equalities or inequalities might we expect for these genomes?
b. Suppose Chargaff had analyzed only the genomes of X174 and B19. What might he have concluded?
c. Suppose Chargaff had included X174 and B19 in his analysis of genomes from other organisms. How might he have altered his conclusions?
a. What base equalities or inequalities might we expect for these genomes?
b. Suppose Chargaff had analyzed only the genomes of X174 and B19. What might he have concluded?
c. Suppose Chargaff had included X174 and B19 in his analysis of genomes from other organisms. How might he have altered his conclusions?

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
22
Different forms of DNA have been identified through X-ray crystallography analysis. These forms include A-DNA, B-DNA, and Z-DNA, and each has unique molecular attributes.
a. What are the molecular attributes of each of these forms of crystallized DNA?
b. Which form is closest in structure to most of the DNA found in living cells? Why isn't cellular DNA identical to this form of crystallized DNA?
c. When, if ever, does cellular DNA have one of the other two forms? What do you infer from this information about the potential cellular role(s) of the other DNA forms?
a. What are the molecular attributes of each of these forms of crystallized DNA?
b. Which form is closest in structure to most of the DNA found in living cells? Why isn't cellular DNA identical to this form of crystallized DNA?
c. When, if ever, does cellular DNA have one of the other two forms? What do you infer from this information about the potential cellular role(s) of the other DNA forms?

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
23
If a virus particle contains 200,000 bp of doublestranded DNA, how many complete 360° turns occur in its genome? (Use the value of 10 bp per turn in your calculation.)

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
24
A double-stranded DNA molecule is 100,000 bp (100 kb) long.
a. How many nucleotides does it contain?
b. How many complete turns are there in the molecule? (Use the value of 10 bp per turn in your calculation.)c. How many nm long is the DNA molecule? (1 nm = 1 × 10 -9 m)
a. How many nucleotides does it contain?
b. How many complete turns are there in the molecule? (Use the value of 10 bp per turn in your calculation.)c. How many nm long is the DNA molecule? (1 nm = 1 × 10 -9 m)

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
25
The bacteriophage T4 genome is 168,900 bp long.
a. What are the dimensions of the genome (in nm) if the molecule remains unfolded as a linear segment of double-stranded DNA?
b. If the T4 protein capsid has about the same dimensions as the capsid of bacteriophage T2 (see Figure), and the thickness of the capsid is about 10 nm, about how many times must the T4 genome be folded to fit into the space available within its capsid?
Figure
Electron micrograph and diagram of bacteriophage T2 (1 nm = 10 -9 m).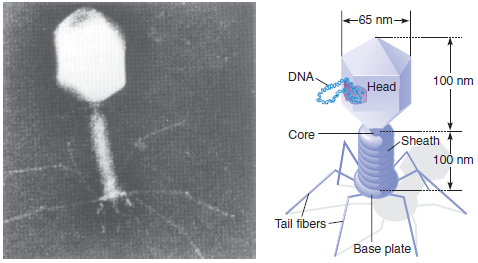
a. What are the dimensions of the genome (in nm) if the molecule remains unfolded as a linear segment of double-stranded DNA?
b. If the T4 protein capsid has about the same dimensions as the capsid of bacteriophage T2 (see Figure), and the thickness of the capsid is about 10 nm, about how many times must the T4 genome be folded to fit into the space available within its capsid?
Figure
Electron micrograph and diagram of bacteriophage T2 (1 nm = 10 -9 m).


Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
26
Different cellular organisms have vastly different amounts of genetic material. E. coli has about 4.6 × 10 6 bp of DNA in one circular chromosome, the haploid budding yeast ( S. cerevisiae ) has 12,067,280 bp of DNA in 16 chromosomes, and the gametes of humans have about 2.75 × 10 9 bp of DNA iN23 chromosomes.
a. For each of these organism's cells, if all of the DNA were B-DNA, what would be the average length of a chromosome in the cell?
b. On average, how many complete turns would be in each chromosome?
c. Would your answers to (a) and (b) be significantly different if the DNA were composed of, say, 20% Z-DNA and 80% B-DNA?
d. What implications do your answers to these questions have for the packaging of DNA in cells?
a. For each of these organism's cells, if all of the DNA were B-DNA, what would be the average length of a chromosome in the cell?
b. On average, how many complete turns would be in each chromosome?
c. Would your answers to (a) and (b) be significantly different if the DNA were composed of, say, 20% Z-DNA and 80% B-DNA?
d. What implications do your answers to these questions have for the packaging of DNA in cells?

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
27
If nucleotides were arranged at random in a piece of single-stranded RNA 10 6 nucleotides long, and if the base composition of this RNA was 20% A , 25% C , 25% U , and 30% G , how many times would you expect the specific sequence 5 ' -GUUA-3 ' to occur?

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
28
Two double-stranded DNA molecules from a population of T2 phages were denatured to single strands by heat treatment. The result was the following four singlestranded DNAs: 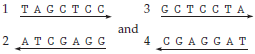
These separated strands were then allowed to renature. Diagram the structures of the renatured molecules most likely to appear when (a) strand 2 renatures with strand 3 and (b) strand 3 renatures with strand 4. Label the strands, and indicate sequences and polarity.
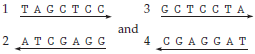
These separated strands were then allowed to renature. Diagram the structures of the renatured molecules most likely to appear when (a) strand 2 renatures with strand 3 and (b) strand 3 renatures with strand 4. Label the strands, and indicate sequences and polarity.

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
29
Define topoisomerases, and list the functions of these enzymes.

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
30
What is the relationship between cellular DNA content and the structural or organizational complexity of the organism?

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
31
Impressive technologies have been developed to sequence entire genomes. Some biotechnology innovators even envision low-cost ($1,000) sequencing of individual human genomes. Still, the genome of the single-celled Ameoba proteus might present a challenge since it has nearly 100 times the DNA content of the human genome (see Table 2.3). If we sequenced its genome, do you expect we would identify about 100-fold more genes than have been found in the human genome? Why or why not? If not, what do you expect we would learn about its genome?
Table Haploid DNA Content, or C-Value, of Selected Species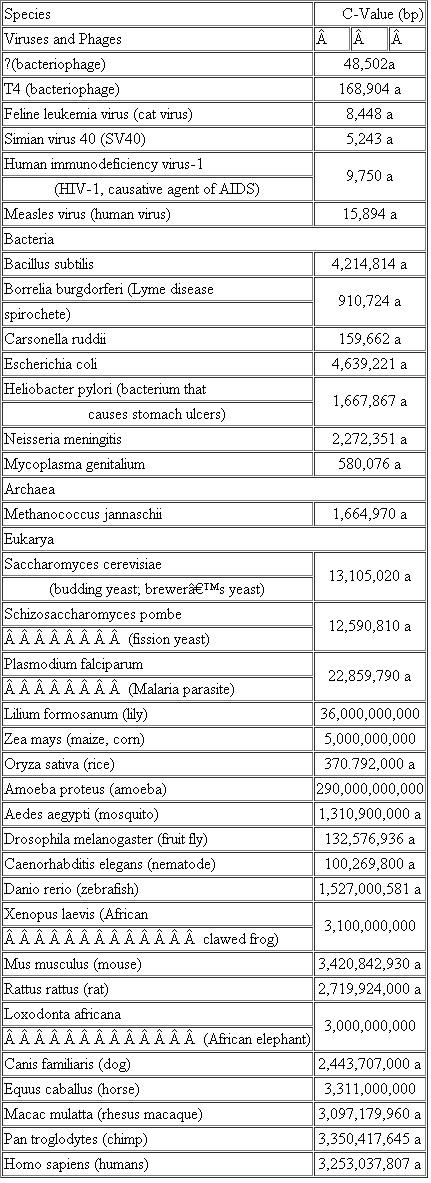
*These C-values derive from the complete genome sequence; all others are estimates based on other measurements.
Table Haploid DNA Content, or C-Value, of Selected Species

*These C-values derive from the complete genome sequence; all others are estimates based on other measurements.

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
32
In a particular eukaryotic chromosome (choose the best answer),
a. heterochromatin and euchromatin are regions where genes make functional gene products (that is, where genes are active).
b. heterochromatin is active, but euchromatin is inactive.
c. heterochromatin is inactive, but euchromatin is active.
d. both heterochromatin and euchromatin are inactive.
a. heterochromatin and euchromatin are regions where genes make functional gene products (that is, where genes are active).
b. heterochromatin is active, but euchromatin is inactive.
c. heterochromatin is inactive, but euchromatin is active.
d. both heterochromatin and euchromatin are inactive.

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
33
Compare and contrast eukaryotic chromosomes and bacterial chromosomes with respect to the following features:
a. centromeres
b. pentose sugars
c. amino acids
d. supercoiling
e. telomeres
f. nonhistone protein scaffolds
g. DNA
h. nucleosomes
i. circular chromosome
j. looping
a. centromeres
b. pentose sugars
c. amino acids
d. supercoiling
e. telomeres
f. nonhistone protein scaffolds
g. DNA
h. nucleosomes
i. circular chromosome
j. looping

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
34
Discuss the components and structure of a nucleosome and the composition of a nucleosome core particle. Explain how nucleosomes are used to package DNA hierarchically.

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
35
Histone proteins from many different eukaryotes are highly similar in their amino acid sequence, making them among the most highly conserved eukaryotic proteins. What functional properties of histone proteins might limit their diversity?

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
36
Set up the following "rope trick": Start with a belt (representing a DNA molecule; imagine the phosphodiester backbones lying along the top and bottom edges of the belt) and a soda can. Holding the belt buckle at the bottom of the can, wrap the belt flat against the side of the can, and wind, counterclockwise three times around the can. Now remove the "core" soda can, and, holding the ends of the belt, pull the ends of the belt taut. After some reflection, answer the following questions:
a. Did you make a left- or a right-handed helix?
b. How many helical turns were present in the coiled belt before it was pulled taut?
c. How many helical turns were present in the coiled belt after it was pulled taut?
d. Why does the belt appear more twisted when pulled taut?
e. About what percentage of the length of the belt was decreased by this packaging?
f. Is the DNA of a linear chromosome that is coiled around histones supercoiled?
g. Why are topoisomerases necessary to package linear chromosomes?
a. Did you make a left- or a right-handed helix?
b. How many helical turns were present in the coiled belt before it was pulled taut?
c. How many helical turns were present in the coiled belt after it was pulled taut?
d. Why does the belt appear more twisted when pulled taut?
e. About what percentage of the length of the belt was decreased by this packaging?
f. Is the DNA of a linear chromosome that is coiled around histones supercoiled?
g. Why are topoisomerases necessary to package linear chromosomes?

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
37
What are the main molecular features of yeast centromeres?

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
38
Telomeres are unique repeated sequences. Where on the DNA strand are they found? Do they serve a function?

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
39
Would you expect to find most protein-coding genes in unique-sequence DNA, in moderately repetitive DNA, or in highly repetitive DNA?

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
40
Both histone and nonhistone proteins are essential for DNA packaging in eukaryotic cells. However, these classes of proteins are fundamentally dissimilar in a number of ways. Describe how they differ in terms of
a. their protein characteristics.
b. their presence and abundance in cells.
c. their interactions with DNA.
d. their role in DNA packaging and the formation of looped domains.
a. their protein characteristics.
b. their presence and abundance in cells.
c. their interactions with DNA.
d. their role in DNA packaging and the formation of looped domains.

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
41
In higher eukaryotes, what relationships exist between these elements?
a. centromeres and tandemly repeated DNA
b. constitutive heterochromatin and centromeric regions
c. euchromatin, facultative heterochromatin, constitutive heterochromatin and unique-sequence DNA
a. centromeres and tandemly repeated DNA
b. constitutive heterochromatin and centromeric regions
c. euchromatin, facultative heterochromatin, constitutive heterochromatin and unique-sequence DNA

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
42
Distinguish between LINEs and SINEs with respect to
a. their length.
b. their abundance in different higher eukaryotic genomes.
c. whether and how they are able to move within a genome.
d. their distribution within a genome.
a. their length.
b. their abundance in different higher eukaryotic genomes.
c. whether and how they are able to move within a genome.
d. their distribution within a genome.

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck
43
Chromosomal rearrangements at the end of 16p (the short arm of chromosome 16) underlie a variety of common human genetic disorders, including
thalassemia (a defect in hemoglobin metabolism caused by mutations in the
globin gene that lies in this region), mental retardation, and the adult form of polycystic kidney disease. Analysis of approximately 285-kb pairs of DNA sequence at the end of human chromosome 16p has allowed for a detailed understanding of the structure of this chromosome region. The first functional gene lies about 44 kb from the region of simple telomeric sequences and about 8 kb from the telomere-associated sequences. Analysis of sequences proximal (nearer the centromere) to the first gene reveals a sinusoidal variation in GC content, with GC -rich regions associated with gene-rich areas and AT -rich regions associated with Alu dense areas. The
globin gene lies about 130 kb from the telomere-associated sequences.
a. Diagram the features of the 16p telomere, and relate them to the current view of telomere structure and function as presented in the text.
b. What have the preceding data revealed about the distribution of SINEs in the terminus of 16p? (SINEs and LINEs are, respectively, short and long interspersed nuclear elements.)
thalassemia (a defect in hemoglobin metabolism caused by mutations in the
globin gene that lies in this region), mental retardation, and the adult form of polycystic kidney disease. Analysis of approximately 285-kb pairs of DNA sequence at the end of human chromosome 16p has allowed for a detailed understanding of the structure of this chromosome region. The first functional gene lies about 44 kb from the region of simple telomeric sequences and about 8 kb from the telomere-associated sequences. Analysis of sequences proximal (nearer the centromere) to the first gene reveals a sinusoidal variation in GC content, with GC -rich regions associated with gene-rich areas and AT -rich regions associated with Alu dense areas. The
globin gene lies about 130 kb from the telomere-associated sequences.
a. Diagram the features of the 16p telomere, and relate them to the current view of telomere structure and function as presented in the text.
b. What have the preceding data revealed about the distribution of SINEs in the terminus of 16p? (SINEs and LINEs are, respectively, short and long interspersed nuclear elements.)

Unlock Deck
Unlock for access to all 43 flashcards in this deck.
Unlock Deck
k this deck


