Deck 20: Nucleic Acids
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/58
Play
Full screen (f)
Deck 20: Nucleic Acids
1
Messenger ribonucleic acid (mRNA) is a single RNA strand encoding a "blueprint" for an amino acid sequence (e.g. a protein, after folding and posttranslational modification). The brief existence of an mRNA macromolecule begins with transcription and ends in enzymatic hydrolysis to ribonucleotides. Whereas mRNA is basically "ready to use" after transcription in non-eukaryotes, eukaryotic mRNA requires extensive processing. In the process called 5'cap addition, a modified 7-methylguanosine ribonucleotide is added via an unusual 5' to 5' triphosphate linkage. What is the chemical structure of the 5'-end of the mRNA strand after capping?
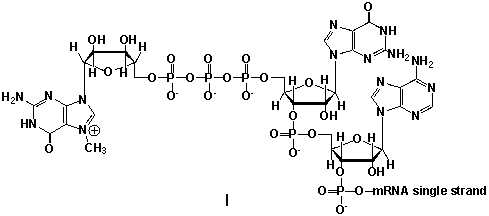
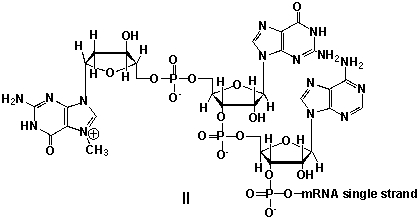
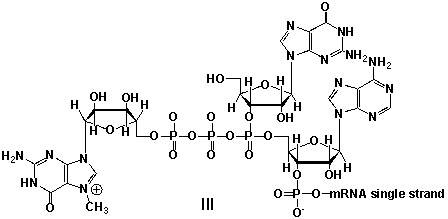
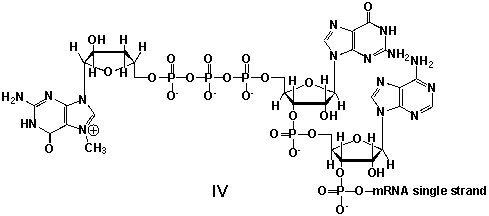
A) I
B) II
C) III
D) IV




A) I
B) II
C) III
D) IV
I
2
Which is the complementary tetranucleotide for 5'-AGCT-3'?
A) 5'-TCGA-3'
B) 5'-CTAG-3'
C) 5'-AGCT-3'
D) 5'-GATC-3'
A) 5'-TCGA-3'
B) 5'-CTAG-3'
C) 5'-AGCT-3'
D) 5'-GATC-3'
5'-AGCT-3'
3
Which is the DNA complement for 5'-ACCGTTAAT-3'?
A) 5'-ATTAACGGT-3'
B) 5'-GTTACCGGC-3'
C) 5'-TGGCAATTA-3'
D) 5'-CAATGGCCG-3'
A) 5'-ATTAACGGT-3'
B) 5'-GTTACCGGC-3'
C) 5'-TGGCAATTA-3'
D) 5'-CAATGGCCG-3'
5'-ATTAACGGT-3'
4
Which are types of RNA?
I. ribosomal
II. histonal
III. helix
IV. transfer
A) I, II
B) I, III
C) II, IV
D) I, IV
I. ribosomal
II. histonal
III. helix
IV. transfer
A) I, II
B) I, III
C) II, IV
D) I, IV

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
5
Which statement about the base mole-percent composition of DNA is true?
A) A=G=C=T
B) (A+G)=(C+T)
C) (A+T)=(G+C)
D) There is no relationship
A) A=G=C=T
B) (A+G)=(C+T)
C) (A+T)=(G+C)
D) There is no relationship

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
6
Which structure is a nucleoside?
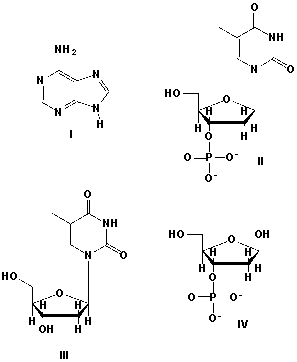
A) I
B) II
C) III
D) IV

A) I
B) II
C) III
D) IV

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
7
Hydrogen bonding is strongest between which two bases?
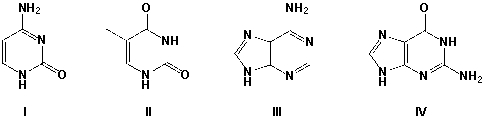
A) I, II
B) I, IV
C) II, III
D) II, IV

A) I, II
B) I, IV
C) II, III
D) II, IV

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
8
DNA-supercoiling occurs because of the following phenomena:
I. Topoisomerases either add to or subtract from DNA-twists.
II. Helicases cause strain when separating the two DNA-single strands that form a DNA double-helix.
III. DNA-double strands are densely packed within chromosomes. Therefore, the twist imposed on one DNA-double strand can cause cooperative supercoiling of the neighboring strands.
IV. RNA-polymerases, the group of enzymes responsible for RNA-transcription can cause supercoiling.
A) I and II
B) II and III
C) I, II, and IV
D) all of the above
I. Topoisomerases either add to or subtract from DNA-twists.
II. Helicases cause strain when separating the two DNA-single strands that form a DNA double-helix.
III. DNA-double strands are densely packed within chromosomes. Therefore, the twist imposed on one DNA-double strand can cause cooperative supercoiling of the neighboring strands.
IV. RNA-polymerases, the group of enzymes responsible for RNA-transcription can cause supercoiling.
A) I and II
B) II and III
C) I, II, and IV
D) all of the above

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
9
Chlorambucil is a chemotherapeutic drug used in the treatment of chronic lymphatic leukemia. It is a nitrogen mustard alkylating agent that can be given orally. The name "nitrogen mustards" stems from the group of alkylating military agents designed for chemical warfare that feature the N-(CH2)2-Cl motif. Which DNA-base can be alkylated easily?(Hint: Which aromatic system has the highest electron density?)
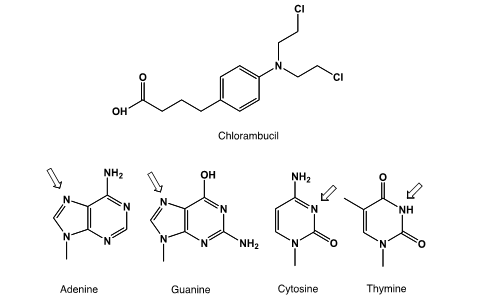
A)Adenine
B)Guanine
C)Cytosine
D)Thymine

A)Adenine
B)Guanine
C)Cytosine
D)Thymine

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
10
Dideoxynucleotides (ddNTPs, shown here is dideoxyguanosine) are nucleosides lacking the 3'-hydroxyl group on their deoxyribose sugar. They are the key molecules for the method of DNA-sequencing, which was developed by Frederick Sanger in 1977. What is the function of dideoxynucleotides, based on its structure?
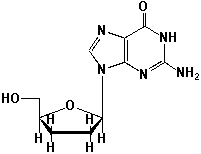
A) Dideoxynucleotides terminate DNA growth during replication at one distinct position.
B) Dideoxynucleotides terminate DNA growth statistically at each A-, G-, C- or T-site, depending on the dideoxynucleotides used (ddA, ddG, ddC or ddT).
C) Dideoxynucleotides terminate the DNA replication and switch on DNA transcription to mRNA.
D) Dideoxynucleotides activate rRNA.

A) Dideoxynucleotides terminate DNA growth during replication at one distinct position.
B) Dideoxynucleotides terminate DNA growth statistically at each A-, G-, C- or T-site, depending on the dideoxynucleotides used (ddA, ddG, ddC or ddT).
C) Dideoxynucleotides terminate the DNA replication and switch on DNA transcription to mRNA.
D) Dideoxynucleotides activate rRNA.

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
11
What is the correct name for this structure?
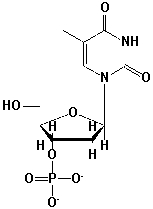
A) 5'-dAMP
B) 5'-TMP
C) 3'-AMP
D) 3'-dTMP

A) 5'-dAMP
B) 5'-TMP
C) 3'-AMP
D) 3'-dTMP

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
12
Which statements are true?
I. A-DNA forms a right-handed helix
II. B-DNA forms a left-handed helix
III. Z-DNA forms a left-handed helix
IV. ribose is a hexose
A) I and II
B) II and III
C) III and IV
D) I and III
I. A-DNA forms a right-handed helix
II. B-DNA forms a left-handed helix
III. Z-DNA forms a left-handed helix
IV. ribose is a hexose
A) I and II
B) II and III
C) III and IV
D) I and III

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
13
Which structures are named correctly?
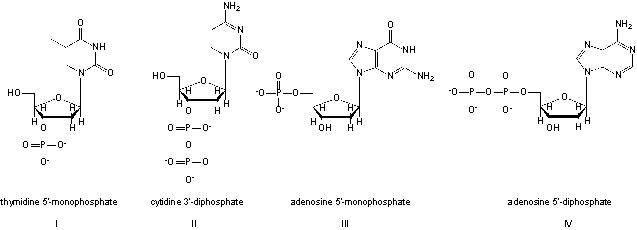
A) I, II
B) II, III
C) I, IV
D) II, IV

A) I, II
B) II, III
C) I, IV
D) II, IV

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
14
Which nucleotides are named correctly?
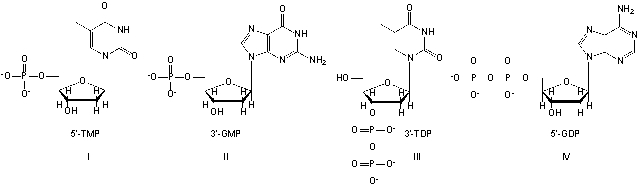
A) I, III
B) II, III
C) III, IV
D) II, IV

A) I, III
B) II, III
C) III, IV
D) II, IV

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
15
A complete turn of the A-DNA helix occurs at which distance?
A) 33 Å
B) 10 Å
C) 25 Å
D) 4 Å
A) 33 Å
B) 10 Å
C) 25 Å
D) 4 Å

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
16
Which structures are named correctly?
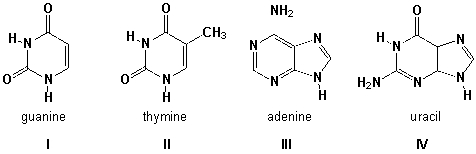
A) I, II
B) II, III
C) III, IV
D) I, III

A) I, II
B) II, III
C) III, IV
D) I, III

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
17
The backbone of DNA is best described as:
A) pairs of complementary bases
B) polyphosphodiesters
C) pyrophosphates
D) sugar glycosides
A) pairs of complementary bases
B) polyphosphodiesters
C) pyrophosphates
D) sugar glycosides

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
18
A complete turn of the B-DNA helix occurs at which distance?
A) 33 Å
B) 10 Å
C) 25 Å
D) 4 Å
A) 33 Å
B) 10 Å
C) 25 Å
D) 4 Å

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
19
Which are the correct pairings of bases in the structure of DNA?
I. A-C
II. G-T
III. A-G
IV. G-C
V. A-T
VI. C-T
A) I, II
B) III, V
C) III, VI
D) IV, V
I. A-C
II. G-T
III. A-G
IV. G-C
V. A-T
VI. C-T
A) I, II
B) III, V
C) III, VI
D) IV, V

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
20
What is the charge on adenosine phosphate at blood serum pH of 7.4?
A) 0
B) -1
C) -2
D) -3
A) 0
B) -1
C) -2
D) -3

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
21
Which is the complementary sequence in RNA for the DNA sequence 5'-AATCAGTT-3'?
A) 5'-AAUCAGUU-3'
B) 5'-AACUGAUU-3'
C) 5'-UUAGUCAA-3'
D) 5'-CCAUCGAA-3'
A) 5'-AAUCAGUU-3'
B) 5'-AACUGAUU-3'
C) 5'-UUAGUCAA-3'
D) 5'-CCAUCGAA-3'

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
22
Because of a mutation the codon for a tetrapeptide: AUGGCAUGGGGGUUGUAG (Ala-Try-Gly-Leu) is changed to AUGGCAUGGGGGUGUAG. What are the consequences for the composition of the tetrapeptide?
A) Ala-Try-Gly
B) Ala-Try-Gly-Leu
C) Ala-Try-Gly-Val
D) Ala-Try-Gly and an undetermined amino acid, because the codon-anticodon binding does not always follow the Watson-Crick pairing rules, especially if the codon/anticodon triplets don't match
A) Ala-Try-Gly
B) Ala-Try-Gly-Leu
C) Ala-Try-Gly-Val
D) Ala-Try-Gly and an undetermined amino acid, because the codon-anticodon binding does not always follow the Watson-Crick pairing rules, especially if the codon/anticodon triplets don't match

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
23
How many nucleotides direct a specific amino acid into a polypeptide sequence?
A) 1
B) 2
C) 3
D) 4
A) 1
B) 2
C) 3
D) 4

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
24
In both RNA and DNA, the phosphate groups are bonded to the sugar structure at which positions?
A) 5', 5'
B) 3', 3'
C) 3', 5'
D) 5', 3'
A) 5', 5'
B) 3', 3'
C) 3', 5'
D) 5', 3'

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
25
Which is the key to in vitro DNA replication?
A) polyacrylamide gels
B) restriction endonucleases
C) primer
D) 2',3'-dideoxynucleoside triphosphate
A) polyacrylamide gels
B) restriction endonucleases
C) primer
D) 2',3'-dideoxynucleoside triphosphate

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
26
Streptozotocin, like other so-called N-nitrosourea derivatives can lead to the oxidation of Cytosine to Uracil. What are the consequences for the complementary nucleobase when the DNA is transcribed?
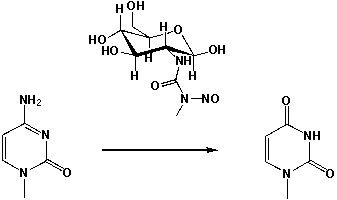
A) No consequence, because thymine and uracil bind both to adenine.
B) In the complementary strand, adenine instead of guanine will bind.
C) In the complementary strand, cytosine instead of guanine will bind.
D) In the complementary strand, guanine instead of adenine will bind.

A) No consequence, because thymine and uracil bind both to adenine.
B) In the complementary strand, adenine instead of guanine will bind.
C) In the complementary strand, cytosine instead of guanine will bind.
D) In the complementary strand, guanine instead of adenine will bind.

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
27
Which is the key to the chain termination method of DNA sequencing?
A) polyacrylamide gels
B) restriction endonucleases
C) primer
D) 2',3'-dideoxynucleoside triphosphate
A) polyacrylamide gels
B) restriction endonucleases
C) primer
D) 2',3'-dideoxynucleoside triphosphate

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
28
Which statements about restriction endonucleases are true?
I. Each cleave DNA at the same site, but leave different fragment lengths.
II. Each recognizes 4 to 8 nucleotides, and cleaves at all sites containing the sequence.
III. Approximately 1000 restriction endonucleases have been isolated and characterized.
IV. Each cleaves the DNA strand by reducing the base off at the sugar 1' position.
A) I, IV
B) II, IV
C) II, III
D) III, IV
I. Each cleave DNA at the same site, but leave different fragment lengths.
II. Each recognizes 4 to 8 nucleotides, and cleaves at all sites containing the sequence.
III. Approximately 1000 restriction endonucleases have been isolated and characterized.
IV. Each cleaves the DNA strand by reducing the base off at the sugar 1' position.
A) I, IV
B) II, IV
C) II, III
D) III, IV

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
29
How do DNA and RNA differ?
I. the position of attachment of phosphate groups
II. the position of attachment of base groups
III. the sugar structure at C-2'
IV. structure of bases
A) I, II
B) II, III
C) III, IV
D) II, IV
I. the position of attachment of phosphate groups
II. the position of attachment of base groups
III. the sugar structure at C-2'
IV. structure of bases
A) I, II
B) II, III
C) III, IV
D) II, IV

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
30
Azidothymidine (AZT) is a nucleoside analog reverse transcriptase inhibitor, which is an antiretroviral drug. It was the first drug approved for the treatment of HIV. AZT was the major breakthrough in AIDS therapy in the 1990's.
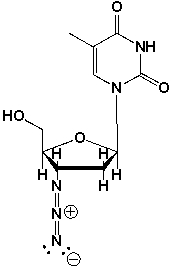
What is the function of AZT, based on its structure?
A) The azido-group (-N3) facilitates irreversible binding to reverse transcriptase.
B) The azido-group (-N3) facilitates irreversible binding to a neighboring nucleotide.
C) AZT cannot be incorporated into a DNA-strand during reverse transcription of the genetic code from RNA to DNA after retroviral infection.
D) All of the above

What is the function of AZT, based on its structure?
A) The azido-group (-N3) facilitates irreversible binding to reverse transcriptase.
B) The azido-group (-N3) facilitates irreversible binding to a neighboring nucleotide.
C) AZT cannot be incorporated into a DNA-strand during reverse transcription of the genetic code from RNA to DNA after retroviral infection.
D) All of the above

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
31
Where does protein synthesis occur?
A) chromatin
B) ribosome
C) codon
D) histones
A) chromatin
B) ribosome
C) codon
D) histones

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
32
Transfer RNA (tRNA) is a small RNA molecule (usually less than 100 nucleotides) that transfers defined amino acids to a growing peptide chain at the ribosomal site during translation. tRNA contains unusual bases, such as, pseudouridine and inosine. Which sequence of ribonucleosides is correct?
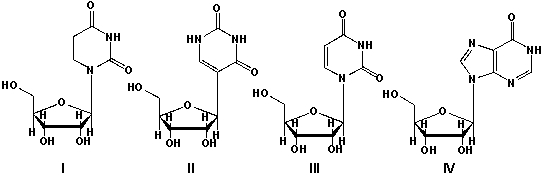
A) I: uridine, II: pseudouridine, III: inosine, IV: dihydrouridine
B) I: pseudouridine, II: uridine, III: inosine, IV:dihydrouridine
C) I: dihydrouridine, II: pseudouridine, III: uridine, IV: inosine
D) I: inosine, II: dihydrouridine, III: uridine, IV:pseudouridine

A) I: uridine, II: pseudouridine, III: inosine, IV: dihydrouridine
B) I: pseudouridine, II: uridine, III: inosine, IV:dihydrouridine
C) I: dihydrouridine, II: pseudouridine, III: uridine, IV: inosine
D) I: inosine, II: dihydrouridine, III: uridine, IV:pseudouridine

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
33
Which is the order of increasing percentage in cells of messenger RNA, transfer RNA, ribosomal RNA (least first)?
A) mRNA, tRNA, rRNA
B) tRNA, rRNA, mRNA
C) rRNA, tRNA, mRNA
D) mRNA, rRNA, tRNA
A) mRNA, tRNA, rRNA
B) tRNA, rRNA, mRNA
C) rRNA, tRNA, mRNA
D) mRNA, rRNA, tRNA

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
34
The name of the following nucleoside is _____________________________________.
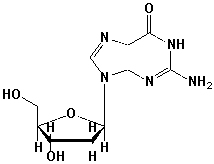


Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
35
Which statements about differences between DNA and RNA are true?
I. The sugar unit is -D-ribose in RNA and -2-deoxyribose in DNA.
II. The pyrimidine bases are uracil and thymine in RNA and cytosine and thymine in DNA.
III. RNA is single stranded, DNA double stranded.
IV. The phosphodiester groups join the 3' to 5' ends of sugars in RNA, and join the 5' to 3' ends
In DNA.
A) I, II
B) I, III
C) II, III
D) III, IV
I. The sugar unit is -D-ribose in RNA and -2-deoxyribose in DNA.
II. The pyrimidine bases are uracil and thymine in RNA and cytosine and thymine in DNA.
III. RNA is single stranded, DNA double stranded.
IV. The phosphodiester groups join the 3' to 5' ends of sugars in RNA, and join the 5' to 3' ends
In DNA.
A) I, II
B) I, III
C) II, III
D) III, IV

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
36
Because of a mutation the codon for a tetrapeptide: AUGGCAUGGGGGUUGUAG (Ala-Try-Gly-Leu) is changed to AUGGCAUGGUGGUUGUAG. What are the consequences for the composition of the tetrapeptide?
A) Ala-Leu-Try-Leu
B) Ala-Gly-Gln-Leu
C) Ala-Try-Try-Leu
D) Gln-Ala-Try-Asn
A) Ala-Leu-Try-Leu
B) Ala-Gly-Gln-Leu
C) Ala-Try-Try-Leu
D) Gln-Ala-Try-Asn

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
37
Whereas mRNA is basically "ready to use" after transcription in non-eukaryotes, eukaryotic mRNA requires extensive processing (see question 21). In the process called polyadenylation a poly(A) tail is added to the 3'end of the mRNA strand (approx. 250 adenosine residues). What is the chemical structure of the 3'-end of the mRNA strand after polyadenylation?
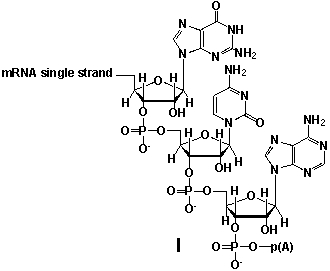
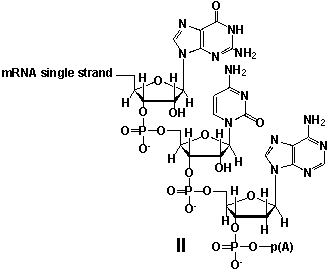
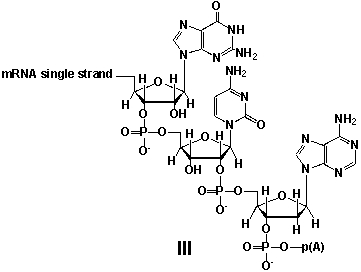
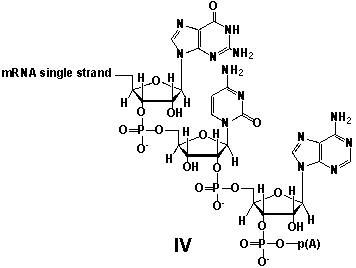
A) I
B) II
C) III
D) IV




A) I
B) II
C) III
D) IV

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
38
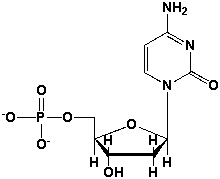
The name of the following nucleotide is _____________________________________.


Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
39
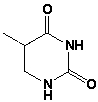
The name of the following base is ____________________________.


Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
40
Which statements about mRNA codons are correct?
I. Each amino acid is coded by only one codon.
II. Codons code for several amino acids.
III. UAA, UAG, UGA are stop codons.
IV. Several amino acids are coded by more than one codon.
A) I, II
B) III, IV
C) I, III
D) II, IV
I. Each amino acid is coded by only one codon.
II. Codons code for several amino acids.
III. UAA, UAG, UGA are stop codons.
IV. Several amino acids are coded by more than one codon.
A) I, II
B) III, IV
C) I, III
D) II, IV

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
41
The complementary DNA single strand of 5'-AGTGTTCCAGT-3' is
5'-____________________________-3'.
5'-____________________________-3'.

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
42
The first four amino acids of the protein coded for by GCUGCCGAUGACGGACGGUGGUGU are __________________________________________________________.

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
43
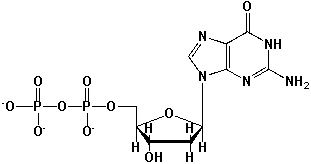
The following nucleotide is named 5'-Guanosine diphosphate.


Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
44
The original DNA strand that results in the following fragments after the Dideoxy method is
5'-_____________________________-3'.
Primer = AGCT
Fragments AAGCT, GAAGCT, AGAAGCT, AAGAAGCT, CAAGAAGCT, TCAAGAAGCT
5'-_____________________________-3'.
Primer = AGCT
Fragments AAGCT, GAAGCT, AGAAGCT, AAGAAGCT, CAAGAAGCT, TCAAGAAGCT

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
45
An mRNA strand that codes for the pentapeptide Met-Ser-Tyr-Met-Cys could have __________ codons in the first position.

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
46
The tertiary structure of DNA involves supercoiling.

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
47
The complementary strand of mRNA to the DNA sequence TAGAAGTAG is ATCTTCATC.

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
48
t-RNA is the lowest molecular weight and most abundant of the RNAs.

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
49
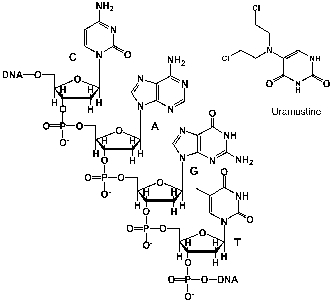
Uramustine or uracil mustard is a chemotherapeutic drug that has been somewhat efficient against lymphatic cancers, such as non-Hodgkin's lymphoma. It works by alkylating the DNA, which causes apoptosis (programmed cell death) of the affected cells. What product is formed during the alkylation of adenine?


Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
50
The Chain Termination method uses dideoxy nucleosides to stop DNA synthesis.

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
51
The complementary RNA strand to the DNA single strand 5'-GGACTTGACTA-3' is
5'-____________________________-3'.
5'-____________________________-3'.

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
52
An mRNA strand that codes for the pentapeptide Ile-Thr-Pro-Lys-Arg could have __________ different codons in the first position.

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
53
The mole % of purine bases must be equal in human DNA.

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
54
The following nucleoside is named Cytidine.
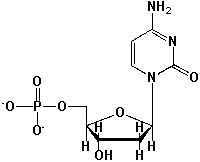


Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
55
If the smallest fragment produced during the Dideoxy method and separated by PAGE includes the primer and the base T, the original DNA template begins with T after the primer sequence.

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
56
mRNA is the shortest lived of the RNAs and the least abundant.

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
57
Streptozotocin is a naturally occurring chemical that is especially toxic to the insulin-producing beta cells of the pancreas in mammals. Streptozotocin is a glucosamine-nitrosourea compound. It is generally alkylating to the DNA bases. What are the structures of all four bases after mono-methylation?
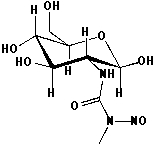
Streptozotocin
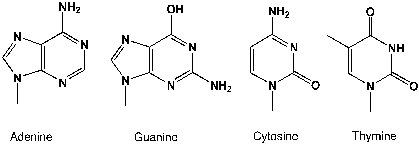

Streptozotocin


Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck
58
tRNA with the codon UGA codes for Cys.

Unlock Deck
Unlock for access to all 58 flashcards in this deck.
Unlock Deck
k this deck


