Deck 6: The Structure of Proteins and Protein: Nucleic Acid Interactions
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/14
Play
Full screen (f)
Deck 6: The Structure of Proteins and Protein: Nucleic Acid Interactions
1
Describe how a ? strand differs from a ? sandwich.
A beta strand is a regular conformation of proteins where the side chains of the amino acids project alternatively to either sides of the backbone. The carbonyl and the amide groups' project laterally and also alternate. The strand is usually seen in a zigzag manner as the backbone is not fully stretched out.
A beta sandwich is a domain where beta sheets are seen. A beta sheet is composed of a group of beta strands. The beta strands are usually seen in opposite arrangement and form a sheet. A beta sandwich has two opposing antiparallel beta sheets. Two groups of beta strands are seen in the form of a sandwich.
A beta sandwich is a domain where beta sheets are seen. A beta sheet is composed of a group of beta strands. The beta strands are usually seen in opposite arrangement and form a sheet. A beta sandwich has two opposing antiparallel beta sheets. Two groups of beta strands are seen in the form of a sandwich.
2
The oxygen-binding proteins hemoglobin and myoglobin differ in that hemoglobin functions as a tetramer in red blood cells, whereas myoglobin functions as a monomer in muscle cells. The globular structure of myoglobin and each hemoglobin monomer involves eight ? -helical segments. Is it the primary, secondary, tertiary, or quaternary structure that differs most between these two proteins? Explain.
Hemoglobin and Myoglobin bind to oxygen in different ways. Hemoglobin functions as a tetramer while Myoglobin functions as a monomer. The structure of myoglobin and one of the units of hemoglobin both have eight alpha helical segments.
The difference between these two proteins is in their quaternary structure. Hemoglobin has a quaternary structure while Myoglobin does not have a quaternary structure. Both these proteins are globular in shape with similar secondary and tertiary structures.
Since the secondary and tertiary structures are similar, we can assume that the primary structure is also similar. Hence, the difference in the quaternary structure is the main reason for the difference in function.
The difference between these two proteins is in their quaternary structure. Hemoglobin has a quaternary structure while Myoglobin does not have a quaternary structure. Both these proteins are globular in shape with similar secondary and tertiary structures.
Since the secondary and tertiary structures are similar, we can assume that the primary structure is also similar. Hence, the difference in the quaternary structure is the main reason for the difference in function.
3
For the following amino acids, suggest whether they are more likely to be found buried or exposed in a stably folded protein domain: phenylalanine, arginine, glutamine, methionine. Explain your answers.
Amino acids can either be on the surface or in the interior of the protein. Those amino acids which are on the surface are usually hydrophilic while those amino acids which are in the interior of the molecule are usually hydrophobiC.• Phenyl alanine - hydrophobic
• Arginine - hydrophilic
• Glutamine - hydrophilic
• Methionine - hydrophobic
The exposed amino acids would be hydrophilic and the hydrophobic amino acids would be buried inside the protein. So, Arginine, and glutamine will be exposed and be present on the surface of the protein. Phenyl alanine and methionine will not be exposed and will be in the interior of the molecule.
• Arginine - hydrophilic
• Glutamine - hydrophilic
• Methionine - hydrophobic
The exposed amino acids would be hydrophilic and the hydrophobic amino acids would be buried inside the protein. So, Arginine, and glutamine will be exposed and be present on the surface of the protein. Phenyl alanine and methionine will not be exposed and will be in the interior of the molecule.
4
You treat a protein with the denaturant urea. For each interaction or bond below, state if the interaction or bond is disrupted by the urea treatment.
A. Ionic bonds.
B. Hydrogen bonds.
C. Disulfide bonds.
D. Peptide bonds.
E. van der Waals interactions.
A. Ionic bonds.
B. Hydrogen bonds.
C. Disulfide bonds.
D. Peptide bonds.
E. van der Waals interactions.

Unlock Deck
Unlock for access to all 14 flashcards in this deck.
Unlock Deck
k this deck
5
From what you learned about the structure of DNA in Chapter 4, explain why Gcn4 interacts with DNA in the major groove rather than in the minor groove. Describe the importance of arginines and lysines in the interaction between Gcn4 and DNA.

Unlock Deck
Unlock for access to all 14 flashcards in this deck.
Unlock Deck
k this deck
6
Predict the effect of substituting one or more of the conserved cysteines or histidines in a Cys2His2 zinc finger with alanine. Explain your answer.

Unlock Deck
Unlock for access to all 14 flashcards in this deck.
Unlock Deck
k this deck
7
Describe the unusual features of the interaction of LEF-1 with DNA.

Unlock Deck
Unlock for access to all 14 flashcards in this deck.
Unlock Deck
k this deck
8
How do enzymes enhance the rate of a reaction?

Unlock Deck
Unlock for access to all 14 flashcards in this deck.
Unlock Deck
k this deck
9
Consider a ligand that is structurally similar to the substrate of an enzyme and that binds tightly in the active site, excluding the normal substrate. What is the difference between such a "competitive inhibitor" and an allosteric inhibitor?

Unlock Deck
Unlock for access to all 14 flashcards in this deck.
Unlock Deck
k this deck
10
A translation initiation factor, called Tif3 or eIRF4B, in yeast cells has the following sequence of elements in its polypeptide chain: an amino-terminal domain containing an RNA recognition motif (RRM), a Central segment with a seven-fold repeated sequence rich in basic and acidic amino acid residues, and a carboxy-terminal region with no evident homology to known motifs or domains.
A. Describe the significance of RRMs.
The modularity of the protein suggested the following series of experiments, to analyze the roles of its different parts. Cells with a deletion of the TIF 3 gene do not grow at 37°C?but grow normally at 30°C. By adding a gene that encodes a fragment of the protein, it is possible to assay for complementation-the capacity of that fragment to confer wild-type growth at 37°C. The results of such experiments are shown in the table below, in which ++?+?and ? indicate the degree of growth/complementation.

B. From these data, which region of the protein is required for wild-type growth at 37°C?
A further set of experiments involved an in vitro translation assay, using an extract from the strain lacking the TIF3 gene. The reaction was initiated by adding purified Tif3 protein or one of its truncated forms. The results are in the table below, which shows the percentage of in vitro translation relative to the reaction with full-length Tif3.
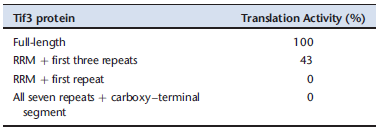
C. How do these results compare with the complementation results in part B? Do they modify your conclusion from the genetic experiments?
Data adapted from Niederberger et al. (1998. RNA 4: 1259-1267).
A. Describe the significance of RRMs.
The modularity of the protein suggested the following series of experiments, to analyze the roles of its different parts. Cells with a deletion of the TIF 3 gene do not grow at 37°C?but grow normally at 30°C. By adding a gene that encodes a fragment of the protein, it is possible to assay for complementation-the capacity of that fragment to confer wild-type growth at 37°C. The results of such experiments are shown in the table below, in which ++?+?and ? indicate the degree of growth/complementation.

B. From these data, which region of the protein is required for wild-type growth at 37°C?
A further set of experiments involved an in vitro translation assay, using an extract from the strain lacking the TIF3 gene. The reaction was initiated by adding purified Tif3 protein or one of its truncated forms. The results are in the table below, which shows the percentage of in vitro translation relative to the reaction with full-length Tif3.

C. How do these results compare with the complementation results in part B? Do they modify your conclusion from the genetic experiments?
Data adapted from Niederberger et al. (1998. RNA 4: 1259-1267).

Unlock Deck
Unlock for access to all 14 flashcards in this deck.
Unlock Deck
k this deck
11
What is the bond that can form between two cys-teines in secreted proteins? Why does this bond not ordinarily form in intracellular proteins? How does this interaction differ from the interactions that can occur between other amino acid side chains?

Unlock Deck
Unlock for access to all 14 flashcards in this deck.
Unlock Deck
k this deck
12
Give an example of two amino acid side chains that can interact with each other through an ionic bond at neutral pH. See Chapter 3 for a review of ionic bonds.

Unlock Deck
Unlock for access to all 14 flashcards in this deck.
Unlock Deck
k this deck
13
A mutation that occurs in DNA can cause an amino acid substitution in the encoded protein. Amino acid substitutions are described as conservative when the amino acid in the mutated protein has chemical properties similar to those of the amino acid it has replaced. Referring to Figure 6-2, identify four different examples of pairs of amino acids that could be involved in conservative substitutions.

Unlock Deck
Unlock for access to all 14 flashcards in this deck.
Unlock Deck
k this deck
14
Peptide bond formation is an example of a condensation reaction. Explain what this statement means and why peptide bond formation is also referred to as a dehydration reaction.

Unlock Deck
Unlock for access to all 14 flashcards in this deck.
Unlock Deck
k this deck


