Deck 20: The Evolution of Genomes
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/47
Play
Full screen (f)
Deck 20: The Evolution of Genomes
1
What can proteomics reveal that genomics cannot?
A) the number of genes characteristic of a species
B) the patterns of alternative splicing
C) the set of proteins present within a cell or tissue type
D) the movement of transposable elements within the genome
A) the number of genes characteristic of a species
B) the patterns of alternative splicing
C) the set of proteins present within a cell or tissue type
D) the movement of transposable elements within the genome
C
2
Which of the following is a direct result of alternative splicing in eukaryotes?
A) More than one polypeptide sequence can be produced from a single gene.
B) Each gene can produce multiple copies of the same peptide.
C) Eukaryote genomes can be smaller than prokaryotic genomes.
D) Introns contain coding sequences and exons do not.
A) More than one polypeptide sequence can be produced from a single gene.
B) Each gene can produce multiple copies of the same peptide.
C) Eukaryote genomes can be smaller than prokaryotic genomes.
D) Introns contain coding sequences and exons do not.
A
3
An indigenous group uses a certain plant for medicinal purposes. Their scientists are most likely to find which of the following in the genome annotation for the medicinal plant?
A) the plant's proteins that are medically important
B) the relative levels of mRNA transcripts produced by the plant
C) the sequences show similarities with known functional domains
D) the cellular location of mRNAs expressed within different plant tissues
A) the plant's proteins that are medically important
B) the relative levels of mRNA transcripts produced by the plant
C) the sequences show similarities with known functional domains
D) the cellular location of mRNAs expressed within different plant tissues
C
4
Which of the following characteristics is unique to multigene families?
A) Multiple genes whose products must be coordinately expressed.
B) Genes whose sequences are very similar and that probably arose by duplication.
C) Genes whose exons can be spliced in a number of different ways.
D) A highly conserved gene found in a number of different species.
A) Multiple genes whose products must be coordinately expressed.
B) Genes whose sequences are very similar and that probably arose by duplication.
C) Genes whose exons can be spliced in a number of different ways.
D) A highly conserved gene found in a number of different species.

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
5
Information within sequence databases, such as GenBank, could be used to perform which of the following tasks?
A) Determine the expression pattern for specific human genes.
B) Construct a tree to determine the evolutionary relationships between various bird species.
C) Search for genes that have not yet been identified in eukaryotic genomes.
D) Compare patterns of gene expression in cancerous and non-cancerous cells.
A) Determine the expression pattern for specific human genes.
B) Construct a tree to determine the evolutionary relationships between various bird species.
C) Search for genes that have not yet been identified in eukaryotic genomes.
D) Compare patterns of gene expression in cancerous and non-cancerous cells.

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
6
Crickets and fruit flies are both insects. Which of the following statements could explain why a certain cricket genome has eleven times as many base pairs as that of the fruit fly, Drosophila melanogaster?
A) Crickets have higher gene density.
B) Drosophila are more complex organisms.
C) Crickets must have more noncoding DNA.
D) Crickets must make many more proteins.
A) Crickets have higher gene density.
B) Drosophila are more complex organisms.
C) Crickets must have more noncoding DNA.
D) Crickets must make many more proteins.

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
7
Using modern techniques of sequencing by synthesis and the shotgun approach, the sequences of entire chromosomes are determined by ________.
A) systematically sequencing DNA sequences as they occur along the length of the chromosome
B) cloning chromosome sections into large plasmid, sequencing, and then reassembling the chromosome
C) fragmenting chromosomal DNA, sequencing and using computer analysis to determine regions where sequences overlap
D) identifying gaps in previously generated genetic maps, cloning those regions into plasmids and determining their DNA sequences
A) systematically sequencing DNA sequences as they occur along the length of the chromosome
B) cloning chromosome sections into large plasmid, sequencing, and then reassembling the chromosome
C) fragmenting chromosomal DNA, sequencing and using computer analysis to determine regions where sequences overlap
D) identifying gaps in previously generated genetic maps, cloning those regions into plasmids and determining their DNA sequences

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
8
Which of the following best describes metagenomics?
A) genome studies that reflect on the effect of environmental conditions on DNA sequences
B) genome studies that compare and annotate representative genes shared by multiple species
C) sequencing DNA from the most highly conserved genes in a lineage
D) sequencing DNA from a community of species from the same ecosystem
A) genome studies that reflect on the effect of environmental conditions on DNA sequences
B) genome studies that compare and annotate representative genes shared by multiple species
C) sequencing DNA from the most highly conserved genes in a lineage
D) sequencing DNA from a community of species from the same ecosystem

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
9
Which of the following is a reason that expressed sequence tags (ESTs) aid the identification of DNA that encodes proteins?
A) ESTs are amino acid sequences that can be used to predict the DNA sequence used in cellular gene expression.
B) ESTs are short DNA sequences conserved among all protein coding regions that can be scanned for by computer software.
C) ESTs are cDNA sequences and since cDNA synthesis uses an mRNA template ESTs identify protein coding genes.
D) ESTs are complementary to DNA promoter sequences and can act as coding region-specific primers.
A) ESTs are amino acid sequences that can be used to predict the DNA sequence used in cellular gene expression.
B) ESTs are short DNA sequences conserved among all protein coding regions that can be scanned for by computer software.
C) ESTs are cDNA sequences and since cDNA synthesis uses an mRNA template ESTs identify protein coding genes.
D) ESTs are complementary to DNA promoter sequences and can act as coding region-specific primers.

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
10
Based on the systems approach employed by the ENCODE project, what percentage of the genome is estimated to be transcribed at some point in at least one cell type?
A) less than 2%
B) about 50%
C) about 75%
D) 100%
A) less than 2%
B) about 50%
C) about 75%
D) 100%

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
11
Which of the following techniques would be most appropriate to test the hypothesis that humans and chimps differ in the expression of a large set of shared genes?
A) RNA-seq
B) polymerase chain reaction (PCR)
C) DNA sequencing
D) protein-protein interaction assays
A) RNA-seq
B) polymerase chain reaction (PCR)
C) DNA sequencing
D) protein-protein interaction assays

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
12
Which of the following statements best defines proteomics?
A) The field working to link each gene to a particular protein.
B) The study of the properties of sets of proteins.
C) The characterization of the functional possibilities of a single protein.
D) The study of how amino acids are ordered in a protein.
A) The field working to link each gene to a particular protein.
B) The study of the properties of sets of proteins.
C) The characterization of the functional possibilities of a single protein.
D) The study of how amino acids are ordered in a protein.

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
13
Which of the following statements is a correct representation of gene density?
A) Humans have 1,000 Mb per genome.
B) C. elegans has ~20,000 genes.
C) Humans have ~20,000 protein-encoding genes in a 3,000 Mb.
D) Saccharomyces has a genome 40 times the size of a human genome.
A) Humans have 1,000 Mb per genome.
B) C. elegans has ~20,000 genes.
C) Humans have ~20,000 protein-encoding genes in a 3,000 Mb.
D) Saccharomyces has a genome 40 times the size of a human genome.

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
14
Which of the following describes an advantage of establishing and using a human reference genome in a bioinformatics investigation?
A) It represents the entire genome of one individual who acts as a standard for comparison.
B) It is continuously revised as new data are collected and represents current consensus.
C) Filling in remaining gaps would be more work than it is worth, so the reference has been set.
D) "Next generation" sequencing efforts can use it to see if their output is correct.
A) It represents the entire genome of one individual who acts as a standard for comparison.
B) It is continuously revised as new data are collected and represents current consensus.
C) Filling in remaining gaps would be more work than it is worth, so the reference has been set.
D) "Next generation" sequencing efforts can use it to see if their output is correct.

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
15
Which of the following processes is part of the whole-genome shotgun approach to sequencing?
A) fragmenting genomic DNA at random sites
B) mapping the chromosomal location of cloned DNA fragments
C) using DNA primers with randomly generated sequences to begin sequencing reactions throughout the genome
D) focusing the efforts to sequence the genomes of new species by using DNA primers known to be conserved between other species
A) fragmenting genomic DNA at random sites
B) mapping the chromosomal location of cloned DNA fragments
C) using DNA primers with randomly generated sequences to begin sequencing reactions throughout the genome
D) focusing the efforts to sequence the genomes of new species by using DNA primers known to be conserved between other species

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
16
A microarray is a tool used in genetic research to determine the relative level of expression of mRNAs in a particular tissue. One use of the technology is in cancer diagnosis and treatment. Which of the following pieces of evidence would support diagnosis of a cancer due to a mutation in a tumor-suppressor gene?
A) The tissue sample shows a high level of gene expression relative to a control (noncancerous) sample.
B) The tissue sample responds to treatment with a mitosis-promoting compound.
C) The mRNAs for the targeted tumor suppressor sequence are not being produced.
D) The mRNAs for cyclins and kinases show unusually high levels of expression.
A) The tissue sample shows a high level of gene expression relative to a control (noncancerous) sample.
B) The tissue sample responds to treatment with a mitosis-promoting compound.
C) The mRNAs for the targeted tumor suppressor sequence are not being produced.
D) The mRNAs for cyclins and kinases show unusually high levels of expression.

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
17
A bioinformatic gene annotation contains which of the following components?
A) a list of predicted transcription start and stop sites and RNA splicing sites in a DNA sequence
B) the name of the gene and a summary its evolutionary connections in other species
C) a description of the function of noncoding regions of the gene
D) a summary of the experimental steps conducted to identify the protein coding region
A) a list of predicted transcription start and stop sites and RNA splicing sites in a DNA sequence
B) the name of the gene and a summary its evolutionary connections in other species
C) a description of the function of noncoding regions of the gene
D) a summary of the experimental steps conducted to identify the protein coding region

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
18
A long region of genomic DNA is compared with a cDNA. The cDNA sequence is identical to segments, but not all, of the genomic DNA sequence. According to these data which of the following statements best describes the genomic DNA?
A) The sequence contains a protein-coding region.
B) The sequence codes for an rRNA.
C) The sequence is an intron.
D) The sequence is a regulatory region.
A) The sequence contains a protein-coding region.
B) The sequence codes for an rRNA.
C) The sequence is an intron.
D) The sequence is a regulatory region.

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
19
Which of the following statements explains the observation that the number of different proteins a vertebrate produces may be larger than the number of genes found within their genome?
A) Vertebrates can produce more than one polypeptide from a single gene.
B) Vertebrate genes contain both exons and introns.
C) Pseudogenes provide alternative regions for translation initiation.
D) Chromosomal rearrangements have increased the number of protein coding regions.
A) Vertebrates can produce more than one polypeptide from a single gene.
B) Vertebrate genes contain both exons and introns.
C) Pseudogenes provide alternative regions for translation initiation.
D) Chromosomal rearrangements have increased the number of protein coding regions.

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
20
Which of the following statements explains why is it more difficult to identify eukaryotic genes than prokaryotic genes using genomic techniques?
A) Proteins are larger in eukaryotes than in prokaryotes.
B) Coding regions of genes in eukaryotes are shorter than in prokaryotes.
C) mRNAs in prokaryotes are usually polycistronic.
D) There are introns in eukaryotic genes.
A) Proteins are larger in eukaryotes than in prokaryotes.
B) Coding regions of genes in eukaryotes are shorter than in prokaryotes.
C) mRNAs in prokaryotes are usually polycistronic.
D) There are introns in eukaryotic genes.

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
21
Fragments of DNA extracted from the remnants of an extinct vertebrate, the woolly mammoth, have been amplified and sequenced. Currently, the most efficient and ethical use of these products is ________.
A) to introduce certain mammoth traits into relatives, such as elephants
B) to clone live woolly mammoths so that their characteristics can be compared with living relatives
C) to examine size and gene density to understand the reasons why mammoths went extinct
D) to better understand the evolutionary relationships among members of related taxa
A) to introduce certain mammoth traits into relatives, such as elephants
B) to clone live woolly mammoths so that their characteristics can be compared with living relatives
C) to examine size and gene density to understand the reasons why mammoths went extinct
D) to better understand the evolutionary relationships among members of related taxa

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
22
Which of the following statements explains how errors during meiosis that result in an extra copy of a chromosome can facilitate evolution?
A) A polyploid individual that results could mate with an individual that is deficient for other chromosomes.
B) Over time, mutations can accumulate in the extra sets of genes and allow evolution of novel functions.
C) The genome is redundant and adding additional copies reinforces the redundancy.
D) Evolution requires fit individuals and an increase in the number of genes in a genome correlates with increased fitness.
A) A polyploid individual that results could mate with an individual that is deficient for other chromosomes.
B) Over time, mutations can accumulate in the extra sets of genes and allow evolution of novel functions.
C) The genome is redundant and adding additional copies reinforces the redundancy.
D) Evolution requires fit individuals and an increase in the number of genes in a genome correlates with increased fitness.

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
23
Which of the following describes a difference between transposable elements and short tandem repeats (STRs)?
A) STRs may result in phenotypic differences between individuals; transposable elements are non-coding regions.
B) STRs occur within introns; transposable elements occur within exons.
C) STRs make up only a small percentage of a given genome while transposable elements often make up larger parts of a given genome.
D) STRs contain much larger repeated units those found in transposable elements.
A) STRs may result in phenotypic differences between individuals; transposable elements are non-coding regions.
B) STRs occur within introns; transposable elements occur within exons.
C) STRs make up only a small percentage of a given genome while transposable elements often make up larger parts of a given genome.
D) STRs contain much larger repeated units those found in transposable elements.

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
24
Which of the following describes an error in a cellular process that results in DNA duplications?
A) exon shuffling
B) alternative splicing
C) incorrect 5 methylation
D) template slippage
A) exon shuffling
B) alternative splicing
C) incorrect 5 methylation
D) template slippage

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
25
In a eukaryote, which of the following describes the most likely outcome when gene duplication occurs in one member of a gene pair?
A) creation of a pseudogene or a gene with a new function
B) creation of an operon that uses common regulatory elements
C) addition of an intron in the original gene
D) nondisjunction of that chromosome during the next round of cell division.
A) creation of a pseudogene or a gene with a new function
B) creation of an operon that uses common regulatory elements
C) addition of an intron in the original gene
D) nondisjunction of that chromosome during the next round of cell division.

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
26
Unequal crossing over during prophase I can result in one sister chromosome with a deletion and another with a duplication. Hemoglobin Lepore is a form of hemoglobin that is missing a series of amino acids. If this form was caused by unequal crossing over, which of the following is the most likely consequence?
A) There should also be individuals whose hemoglobin contains a tandem repeat of the amino acid sequence that was deleted in hemoglobin Lepore.
B) Each of the genes in the hemoglobin gene family must show the same deletion.
C) The gene with the deletion must have undergone exon shuffling.
D) The deleted region must be located in a different area of the individual's genome.
A) There should also be individuals whose hemoglobin contains a tandem repeat of the amino acid sequence that was deleted in hemoglobin Lepore.
B) Each of the genes in the hemoglobin gene family must show the same deletion.
C) The gene with the deletion must have undergone exon shuffling.
D) The deleted region must be located in a different area of the individual's genome.

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
27
Homeotic genes contain a homeobox sequence that is highly conserved among diverse species. The homeobox sequence encodes a protein domain that binds DNA and regulates development. Which of the following statements is correct regarding homeotic genes?
A) Homeotic genes are selectively expressed as an organism develops.
B) Homeoboxes cannot be expressed in invertebrate species.
C) Homeotic genes in apes and humans are very different.
D) Homeobox sequences have shown convergent evolution among species.
A) Homeotic genes are selectively expressed as an organism develops.
B) Homeoboxes cannot be expressed in invertebrate species.
C) Homeotic genes in apes and humans are very different.
D) Homeobox sequences have shown convergent evolution among species.

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
28
Which of the following statements distinguishes between the way transposons and retrotransposons move around in a genome?
A) Only transposons use a transposase to move.
B) Only transposons use a DNA intermediate.
C) Only transposons use an RNA intermediate.
D) Only transposons use the original copy as a template.
A) Only transposons use a transposase to move.
B) Only transposons use a DNA intermediate.
C) Only transposons use an RNA intermediate.
D) Only transposons use the original copy as a template.

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
29
Which of the following processes is most likely responsible for the variable number of repeated units found at a specific STR locus?
A) failure of homologous chromosomes to separate during meiosis
B) mutations causing increased activity in rRNA
C) slippage during DNA replication
D) alternative splicing of transcripts
A) failure of homologous chromosomes to separate during meiosis
B) mutations causing increased activity in rRNA
C) slippage during DNA replication
D) alternative splicing of transcripts

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
30
Which of the following statements explains how hemoglobin in a human embryo has a higher affinity for oxygen than hemoglobin in the adult?
A) Nonidentical genes produce different versions of globins at different stages of development.
B) Globin pseudogenes interfere with gene expression in adults.
C) Imprinting of globin genes changes the type of hemoglobin produced after birth.
D) Throughout development, mutations accumulate during DNA replication altering the adult form.
A) Nonidentical genes produce different versions of globins at different stages of development.
B) Globin pseudogenes interfere with gene expression in adults.
C) Imprinting of globin genes changes the type of hemoglobin produced after birth.
D) Throughout development, mutations accumulate during DNA replication altering the adult form.

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
31
Which of the following statements best describes multi gene families?
A) a set of linked genes that are transcribed as a single unit
B) repeated sequences of genetic material that are inherited
C) a collection of two or more identical or very similar genes
D) related individuals that share more than one phenotypic trait
A) a set of linked genes that are transcribed as a single unit
B) repeated sequences of genetic material that are inherited
C) a collection of two or more identical or very similar genes
D) related individuals that share more than one phenotypic trait

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
32
Exon shuffling occurs during which of the following processes?
A) splicing of DNA
B) DNA replication
C) meiotic recombination
D) transposon "jumping"
A) splicing of DNA
B) DNA replication
C) meiotic recombination
D) transposon "jumping"

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
33
Human genomes contain several globin genes that are expressed at distinct times in development. Which of the mechanisms listed best explains this developmental pattern?
A) exon shuffling
B) pseudogene activation
C) differential translation of mRNAs
D) differential gene regulation over time
A) exon shuffling
B) pseudogene activation
C) differential translation of mRNAs
D) differential gene regulation over time

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
34
Which of the following statements correctly describes retrotransposons?
A) They use an RNA molecule as an intermediate in transposition.
B) They are found only in animal cells.
C) They generally move by a cut-and-paste mechanism.
D) They contribute a significant portion of the genetic variability seen within a population of gametes.
A) They use an RNA molecule as an intermediate in transposition.
B) They are found only in animal cells.
C) They generally move by a cut-and-paste mechanism.
D) They contribute a significant portion of the genetic variability seen within a population of gametes.

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
35
A recent study compared the Homo sapiens genome with that of Neanderthals. The results indicated that the two genomes mixed at some period in evolutionary history. Which of the following data would provide additional support for this hypothesis?
A) finding that Neanderthals and humans have lived in the same geographical areas
B) DNA from a modern H. sapiens that contains some Neanderthal sequences
C) duplications of several Neanderthal genes on a Neanderthal chromosome
D) determining that some Neanderthal chromosomes are shorter than their counterparts in living humans
A) finding that Neanderthals and humans have lived in the same geographical areas
B) DNA from a modern H. sapiens that contains some Neanderthal sequences
C) duplications of several Neanderthal genes on a Neanderthal chromosome
D) determining that some Neanderthal chromosomes are shorter than their counterparts in living humans

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
36
Humans have 23 pairs of chromosomes, and chimps have 24 pairs of chromosomes. Which of the following best explains the difference in chromosome number between humans and chimps?
A) The common ancestor of humans and chimps had 24 pairs of chromosomes. During human evolution, two human chromosomes fused end to end.
B) In the evolution of chimps, new adaptations resulted from additional chromosomal material.
C) At some point in evolution, human and chimp ancestors reproduced with each other.
D) An Error in mitosis in a common ancestor split one chromosome and resulted in the additional pair seen in chimps.
A) The common ancestor of humans and chimps had 24 pairs of chromosomes. During human evolution, two human chromosomes fused end to end.
B) In the evolution of chimps, new adaptations resulted from additional chromosomal material.
C) At some point in evolution, human and chimp ancestors reproduced with each other.
D) An Error in mitosis in a common ancestor split one chromosome and resulted in the additional pair seen in chimps.

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
37
Use the following information to answer the question. 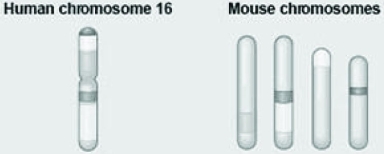 The figure shows a diagram of blocks of genes on human chromosome 16 and the locations of blocks of similar genes as they are found on four different mouse chromosomes.
The figure shows a diagram of blocks of genes on human chromosome 16 and the locations of blocks of similar genes as they are found on four different mouse chromosomes.
Which of the following statements best explains the different arrangements of these blocks in the genomes of these two species?
A) During evolutionary time, these sequences separated and have returned to their original positions in the mouse.
B) DNA sequences within these blocks have become increasingly divergent.
C) Sequences represented have duplicated at least three times resulting in this arrangement.
D) Chromosomal translocations have moved blocks of sequences to other chromosomes.
 The figure shows a diagram of blocks of genes on human chromosome 16 and the locations of blocks of similar genes as they are found on four different mouse chromosomes.
The figure shows a diagram of blocks of genes on human chromosome 16 and the locations of blocks of similar genes as they are found on four different mouse chromosomes.Which of the following statements best explains the different arrangements of these blocks in the genomes of these two species?
A) During evolutionary time, these sequences separated and have returned to their original positions in the mouse.
B) DNA sequences within these blocks have become increasingly divergent.
C) Sequences represented have duplicated at least three times resulting in this arrangement.
D) Chromosomal translocations have moved blocks of sequences to other chromosomes.

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
38
Which of the following best explains the presence of homologous genes in different species?
A) the species descended from a common ancestor
B) the homology developed due to convergent evolution
C) chance mutations were acted on by regional environmental selective pressures
D) analogous structures require proteins encoded by homologous genes
A) the species descended from a common ancestor
B) the homology developed due to convergent evolution
C) chance mutations were acted on by regional environmental selective pressures
D) analogous structures require proteins encoded by homologous genes

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
39
Which of the following statements best explains why it is more difficult to determine a complete eukaryotic genome sequence than it is to determine the complete genomes of bacteria or archaea?
A) Eukaryotic proteins are larger in size are encoded by longer genes.
B) Eukaryotic genomes contain sequences for hard-to-find proteins.
C) Eukaryotic DNA has a high proportion of G-C base pairs which makes sequencing difficult to complete.
D) Eukaryotes have a large amount of repetitive DNA making sequence alignment difficult.
A) Eukaryotic proteins are larger in size are encoded by longer genes.
B) Eukaryotic genomes contain sequences for hard-to-find proteins.
C) Eukaryotic DNA has a high proportion of G-C base pairs which makes sequencing difficult to complete.
D) Eukaryotes have a large amount of repetitive DNA making sequence alignment difficult.

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
40
If gene duplication occurs to its ultimate extent and doubles all genes in a genome, which of the following outcomes is most likely to occur?
A) creation of a pseudogene
B) creation of a gene cluster
C) creation of a polyploid
D) creation of a diploid
A) creation of a pseudogene
B) creation of a gene cluster
C) creation of a polyploid
D) creation of a diploid

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
41
Which of the following types of genome variations are more likely to have phenotypic consequences?
A) CNVs
B) SNPs
C) STRs
D) Alu elements
A) CNVs
B) SNPs
C) STRs
D) Alu elements

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
42
Studies in knockout mice have demonstrated an important role of the FOXP2 transcription factor in the development of vocalizations. Recent sequence comparisons of the FOXP2 gene in Neanderthals and modern humans show that while the DNA sequence may be different, the protein sequence it codes for is identical. Which of the following conclusions might logically be inferred from this information?
A) Because different DNA sequences cannot result in the same protein sequence Neanderthals must have used a different variant of the genetic code.
B) Because there were differences in DNA sequence the hypothesis that Neanderthals were primitive beings that had not developed language is supported.
C) Because the FOXP2 protein sequences were identical, human and Neanderthal vocalizations may have been more similar than previously thought.
D) Because mice containing the "humanized" FOXP2 gene vocalized differently, mice and humans likely diverged more recently than previously thought.
A) Because different DNA sequences cannot result in the same protein sequence Neanderthals must have used a different variant of the genetic code.
B) Because there were differences in DNA sequence the hypothesis that Neanderthals were primitive beings that had not developed language is supported.
C) Because the FOXP2 protein sequences were identical, human and Neanderthal vocalizations may have been more similar than previously thought.
D) Because mice containing the "humanized" FOXP2 gene vocalized differently, mice and humans likely diverged more recently than previously thought.

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
43
Bioinformatics includes all of the following except ________.
A) using computer programs to align DNA sequences
B) using DNA technology to combine DNA from two different sources in a test tube
C) developing computer-based tools for genome analysis
D) using mathematical tools to make sense of biological systems
A) using computer programs to align DNA sequences
B) using DNA technology to combine DNA from two different sources in a test tube
C) developing computer-based tools for genome analysis
D) using mathematical tools to make sense of biological systems

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
44
Two eukaryotic proteins have one domain in common but are otherwise very different. Which of the following processes is most likely to have contributed to this similarity?
A) gene duplication
B) alternative splicing
C) exon shuffling
D) random point mutations
A) gene duplication
B) alternative splicing
C) exon shuffling
D) random point mutations

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
45
Comparisons of DNA sequences within the human species have revealed many variations. Which of the following variations involves duplication of relatively long stretches of DNA?
A) CNVs
B) SNPs
C) STRs
D) Transposable elements
A) CNVs
B) SNPs
C) STRs
D) Transposable elements

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
46
Humans and chimpanzees show approximately 98% sequence similarity yet exhibit significant phenotypic differences. Changes to which of the following genome characteristics contributes most to the differences between humans and chimpanzees?
A) structural genes
B) the number of repeated sequences
C) regulatory sequences
D) genome size
A) structural genes
B) the number of repeated sequences
C) regulatory sequences
D) genome size

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck
47
Homeotic genes ________.
A) encode transcription factors that control the expression of genes responsible for specific anatomical structures
B) are found only in Drosophila and other arthropods
C) are the only genes that contain the homeobox domain
D) encode proteins that form anatomical structures in the fly
A) encode transcription factors that control the expression of genes responsible for specific anatomical structures
B) are found only in Drosophila and other arthropods
C) are the only genes that contain the homeobox domain
D) encode proteins that form anatomical structures in the fly

Unlock Deck
Unlock for access to all 47 flashcards in this deck.
Unlock Deck
k this deck


