Deck 5: DNA Replication, Repair, and Recombination
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/51
Play
Full screen (f)
Deck 5: DNA Replication, Repair, and Recombination
1
What do the enzymes topoisomerase I and topoisomerase II have in common?
A) They both have nuclease activity.
B) They both create double-strand DNA breaks.
C) They both require ATP hydrolysis for their function.
D) They both can create winding (tension) in an initially relaxed DNA molecule.
E) All of the above.
A) They both have nuclease activity.
B) They both create double-strand DNA breaks.
C) They both require ATP hydrolysis for their function.
D) They both can create winding (tension) in an initially relaxed DNA molecule.
E) All of the above.
A
Explanation: Unlike topoisomerase I, which can only relieve the tension in DNA through introducing a nick (single-strand break) in one of the DNA strands, topoisomerase II can actively introduce or relieve tension by creating double-strand breaks (that remain tightly associated with the enzyme) using energy from ATP hydrolysis.
Explanation: Unlike topoisomerase I, which can only relieve the tension in DNA through introducing a nick (single-strand break) in one of the DNA strands, topoisomerase II can actively introduce or relieve tension by creating double-strand breaks (that remain tightly associated with the enzyme) using energy from ATP hydrolysis.
2
You have found a strain of Escherichia coli that has an unusually short doubling time of only 15 minutes, despite the fact that its complete DNA replication should take almost 35 minutes. You also find that there is only one replication origin on its chromosome from which two forks originate, just like the normal process described for E. coli. However, you discover that the origin of replication in this strain has a significantly shorter "refractory period," resulting in the reactivation of the origin before the previous round of replication is over. Based on this model, if you examine the chromosomes of this strain (under conditions of fast growth), how many replication forks would you expect to observe per chromosome on average?
A) Two, just like the wild-type strain
B) Four
C) Six
D) Eight
E) Ten
A) Two, just like the wild-type strain
B) Four
C) Six
D) Eight
E) Ten
C
Explanation: Even though each round of replication takes 35 minutes, the "firing" interval can be adjusted independently, and even go out of control. In this strain, possibly due to mutations in the methylation pathway, the origins can become activated every 15 minutes, resulting in an average of six forks (three bubbles) per chromosome, the newest of which have started about 30 minutes after the oldest ones.
Explanation: Even though each round of replication takes 35 minutes, the "firing" interval can be adjusted independently, and even go out of control. In this strain, possibly due to mutations in the methylation pathway, the origins can become activated every 15 minutes, resulting in an average of six forks (three bubbles) per chromosome, the newest of which have started about 30 minutes after the oldest ones.
3
The Dam methylase is responsible for methylating the adenine base in GATC sequences in Escherichia coli. Imagine two
A) Both of the strains
B) Neither of them
C) Only the first strain
D) Only the second strain
E) coli strains, one without any active Dam methylase, and the other with a hyperactive version of the enzyme that operates faster than the wild-type enzyme. Which of these strains would you expect to show a "mutator" phenotype?
A) Both of the strains
B) Neither of them
C) Only the first strain
D) Only the second strain
E) coli strains, one without any active Dam methylase, and the other with a hyperactive version of the enzyme that operates faster than the wild-type enzyme. Which of these strains would you expect to show a "mutator" phenotype?
A
Explanation: An inactive or hyperactive Dam methylase can interfere with strand-directed mismatch repair, as both make it harder to distinguish between the newly replicated and old DNA strands. Both situations are expected to increase the overall mutation rate, giving rise to a mutator phenotype.
Explanation: An inactive or hyperactive Dam methylase can interfere with strand-directed mismatch repair, as both make it harder to distinguish between the newly replicated and old DNA strands. Both situations are expected to increase the overall mutation rate, giving rise to a mutator phenotype.
4
This protein is present at every replication fork and prevents DNA polymerase from dissociating, but does not impede the rapid movement of the enzyme. Which of the following is true regarding this protein?
A) It self-assembles onto DNA at the replication fork.
B) It is assembled on DNA as soon as DNA polymerase runs into a double-strand region of DNA.
C) Its assembly normally follows the synthesis of a new primer by the DNA primase.
D) It disassembles from DNA as soon as DNA polymerase runs into a double-strand region.
E) All of the above.
A) It self-assembles onto DNA at the replication fork.
B) It is assembled on DNA as soon as DNA polymerase runs into a double-strand region of DNA.
C) Its assembly normally follows the synthesis of a new primer by the DNA primase.
D) It disassembles from DNA as soon as DNA polymerase runs into a double-strand region.
E) All of the above.

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
5
At the replication fork, the template for the lagging strand is thought to loop around. This looping would allow the lagging-strand polymerase to move along with the rest of the replication fork instead of in the opposite direction. The single-strand part of the loop is bound by the single-strand DNA-binding (SSB) proteins. As each Okazaki fragment is synthesized toward completion, how does the size of the loop change? What about the size of the SSB-bound part of the loop?
A) Increases; increases.
B) Increases; decreases.
C) Decreases; increases.
D) Decreases; decreases.
E) Decreases; does not change.
A) Increases; increases.
B) Increases; decreases.
C) Decreases; increases.
D) Decreases; decreases.
E) Decreases; does not change.

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
6
The telomerase enzyme in human cells …
A) has an RNA component.
B) extends the telomeres by its RNA polymerase activity.
C) polymerizes the telomeric DNA sequences without using any template.
D) removes telomeric DNA from the ends of the chromosomes.
E) creates the "end-replication" problem.
A) has an RNA component.
B) extends the telomeres by its RNA polymerase activity.
C) polymerizes the telomeric DNA sequences without using any template.
D) removes telomeric DNA from the ends of the chromosomes.
E) creates the "end-replication" problem.

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
7
If this protein complex does not function normally, the ends of the eukaryotic chromosomes would activate the cell's DNA damage response, causing chromosomal fusions and other genomic anomalies. What is this protein complex called?
A) Telomerase
B) T-loop
C) ORC
D) Shelterin
E) RecA
A) Telomerase
B) T-loop
C) ORC
D) Shelterin
E) RecA

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
8
Fill in the gap in the following paragraph using what you know about the activities of the proteins involved in DNA replication.
"Mitochondrial DNA replication requires a set of proteins similar to those used for the replication of the nuclear genome. However, mitochondria lack a dedicated DNA ... and use the mitochondrial RNA polymerase instead."
"Mitochondrial DNA replication requires a set of proteins similar to those used for the replication of the nuclear genome. However, mitochondria lack a dedicated DNA ... and use the mitochondrial RNA polymerase instead."

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
9
In Escherichia coli, replication of DNA can occur throughout the cell cycle while the cell is also actively transcribing its genes. This means collisions between replication forks and RNA polymerases are inevitable. Depending on the orientation of the genes, collisions can be rear-end (when both machines are traveling in the same direction) or head-on (when they are traveling in opposite directions). In the following paragraph, match each of the letters (A to D) to one appropriate number below. Do not use a number more than once. Your answer would be a four-digit number composed of digits 1 to 5 only, e.g. 1253.
"Typically, in a rear-end collision, the (A) of RNA polymerase collides with the (B) in the replication fork. In contrast, in a head-on collision, the (C) of RNA polymerase hits the (D) in the fork."
1. front edge (of RNA polymerase)
2. rear edge (of RNA polymerase)
3. DNA helicase
4. leading-strand DNA polymerase
5. lagging-strand DNA polymerase
"Typically, in a rear-end collision, the (A) of RNA polymerase collides with the (B) in the replication fork. In contrast, in a head-on collision, the (C) of RNA polymerase hits the (D) in the fork."
1. front edge (of RNA polymerase)
2. rear edge (of RNA polymerase)
3. DNA helicase
4. leading-strand DNA polymerase
5. lagging-strand DNA polymerase

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
10
On average, errors occur in DNA synthesis only once in every ten billion nucleotides incorporated. Which of the following does NOT contribute to this high fidelity of DNA synthesis?
A) Complementary base-pairing between the nucleotides
B) "Tightening" of the DNA polymerase enzyme around its active site to ensure correct pairing before monomer incorporation
C) Exonucleolytic proofreading by the 3′-to-5′ exonuclease activity of the enzyme to correct mispairing even after monomer incorporation
D) A strand-directed mismatch repair system that detects and resolves mismatches soon after DNA replication
E) All of the above mechanisms DO contribute to the fidelity.
A) Complementary base-pairing between the nucleotides
B) "Tightening" of the DNA polymerase enzyme around its active site to ensure correct pairing before monomer incorporation
C) Exonucleolytic proofreading by the 3′-to-5′ exonuclease activity of the enzyme to correct mispairing even after monomer incorporation
D) A strand-directed mismatch repair system that detects and resolves mismatches soon after DNA replication
E) All of the above mechanisms DO contribute to the fidelity.

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
11
In the following schematic drawing, two DNA molecules are shown before and after the action of a protein that is also involved in the process of DNA replication. What is this protein called? 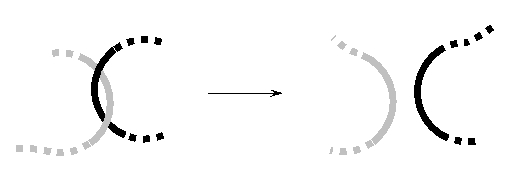
A) DNA ligase
B) DNA helicase
C) DNA polymerase I
D) DNA topoisomerase I
E) DNA topoisomerase II

A) DNA ligase
B) DNA helicase
C) DNA polymerase I
D) DNA topoisomerase I
E) DNA topoisomerase II

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
12
During DNA replication, the single-strand DNA-binding (SSB) proteins …
A) are generally found more on the leading strand than the lagging strand.
B) bind cooperatively to single-stranded DNA and cover the bases to prevent base-pairing.
C) prevent the folding of the single-stranded DNA.
D) bind cooperatively to short hairpin helices that readily form in the single-stranded DNA.
E) All of the above.
A) are generally found more on the leading strand than the lagging strand.
B) bind cooperatively to single-stranded DNA and cover the bases to prevent base-pairing.
C) prevent the folding of the single-stranded DNA.
D) bind cooperatively to short hairpin helices that readily form in the single-stranded DNA.
E) All of the above.

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
13
The mutation rate in bacteria is about 3 nucleotide changes per 10 billion nucleotides per cell generation. Under laboratory conditions, bacteria such as Escherichia coli can divide and double in number about every 40 minutes. If a single Escherichia coli cell is allowed to exponentially divide for 10 hours in this manner, how many mutations would you expect to observe on average in the genome (4.5 million nucleotide pairs) of each of the resulting bacteria compared to the original cell? Assume all mutations are neutral; that is, they do not affect the cell-division time.
A) Less than 0.001
B) About 0.02
C) One or two
D) About 10
E) About 100
A) Less than 0.001
B) About 0.02
C) One or two
D) About 10
E) About 100

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
14
Which of the following features is common between the replication origins in Escherichia coli and Saccharomyces cerevisiae?
A) They both normally exist in one copy per genome.
B) Both are specified by DNA sequences of tens of thousands of nucleotide pairs.
C) Both contain sequences that attract initiator proteins, as well as stretches of DNA rich in A-T base pairs.
D) Both contain GATC repeats that are methylated to prevent the inappropriate "firing" of the origin.
E) All of the above.
A) They both normally exist in one copy per genome.
B) Both are specified by DNA sequences of tens of thousands of nucleotide pairs.
C) Both contain sequences that attract initiator proteins, as well as stretches of DNA rich in A-T base pairs.
D) Both contain GATC repeats that are methylated to prevent the inappropriate "firing" of the origin.
E) All of the above.

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
15
DNA ligases are used in both DNA replication and repair to seal breaks in the DNA. But DNA damage can result in single- or double-strand breaks that are not normal ligase substrates. These need to be processed first before a ligase can act on them. One of the enzymes that is recruited to some of such breaks is called PNK. It has two separate activities on the DNA, both of which can help provide a canonical ligase substrate. Which of the following activities would you expect PNK to have in this context?
A) 5′ kinase (phosphorylation of a free 5′-OH group) and 3′ kinase
B) 5′ phosphatase (dephosphorylation to create a free 5′-OH group) and 3′ phosphatase
C) 3′ kinase and 3′ phosphatase
D) 5′ phosphatase and 3′ kinase
E) 5′ kinase and 3′ phosphatase
A) 5′ kinase (phosphorylation of a free 5′-OH group) and 3′ kinase
B) 5′ phosphatase (dephosphorylation to create a free 5′-OH group) and 3′ phosphatase
C) 3′ kinase and 3′ phosphatase
D) 5′ phosphatase and 3′ kinase
E) 5′ kinase and 3′ phosphatase

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
16
What is the main source of the free energy for the mechanical work performed by DNA helicases during DNA replication in our cells?
A) The hydrogen-bonding energy in the DNA double helix
B) Thermal energy in the nucleus
C) ATP hydrolysis by the helicase
D) The energy of SSB binding to single-stranded DNA
E) ATP hydrolysis by DNA topoisomerases
A) The hydrogen-bonding energy in the DNA double helix
B) Thermal energy in the nucleus
C) ATP hydrolysis by the helicase
D) The energy of SSB binding to single-stranded DNA
E) ATP hydrolysis by DNA topoisomerases

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
17
The nuclear DNA polymerases in human cells …
A) polymerize about 1000 nucleotides per second during DNA replication in vivo.
B) are incapable of 3′-to-5′ exonuclease activity.
C) are capable of 3′-to-5′ DNA polymerase activity.
D) have a single active site that is used for both polymerization and editing.
E) are unable to initiate polymerization de novo (i.e. in the absence of a primer).
A) polymerize about 1000 nucleotides per second during DNA replication in vivo.
B) are incapable of 3′-to-5′ exonuclease activity.
C) are capable of 3′-to-5′ DNA polymerase activity.
D) have a single active site that is used for both polymerization and editing.
E) are unable to initiate polymerization de novo (i.e. in the absence of a primer).

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
18
Which of the following is correct regarding the mutation rate of genomic DNA in different organisms?
A) Human cells have a much higher mutation rate compared to bacteria when the rate is normalized to a single round of replication over the same length of DNA.
B) Mutation rates limit the number of essential genes in an organism's genome.
C) Mutations in the somatic cells cannot be lethal.
D) Even if the mutation rate was 10 times higher than its current value, germ-cell stability in humans would not have been affected.
E) All of the above.
A) Human cells have a much higher mutation rate compared to bacteria when the rate is normalized to a single round of replication over the same length of DNA.
B) Mutation rates limit the number of essential genes in an organism's genome.
C) Mutations in the somatic cells cannot be lethal.
D) Even if the mutation rate was 10 times higher than its current value, germ-cell stability in humans would not have been affected.
E) All of the above.

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
19
During DNA replication in the cell, DNA primase makes short primers that are then extended by the replicative DNA polymerases. These primers …
A) are made up of DNA.
B) generally have a higher number of mutations compared to their neighboring DNA.
C) are made more frequently in the leading strand than the lagging strand.
D) are joined to the neighboring DNA by DNA ligase.
E) provide a 3′-phosphate group for the DNA polymerases to extend.
A) are made up of DNA.
B) generally have a higher number of mutations compared to their neighboring DNA.
C) are made more frequently in the leading strand than the lagging strand.
D) are joined to the neighboring DNA by DNA ligase.
E) provide a 3′-phosphate group for the DNA polymerases to extend.

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
20
Which of the following schematic drawings better depicts the end of mammalian chromosomal DNA? 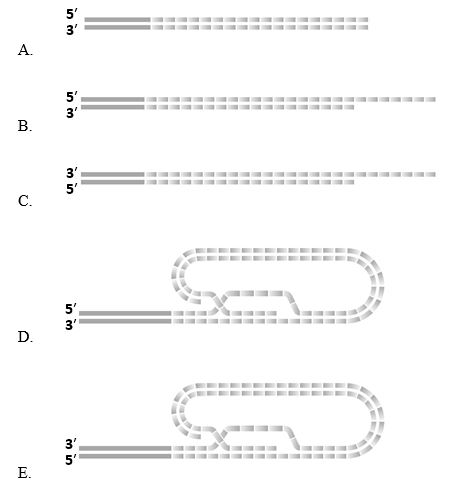


Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
21
In the following schematic drawing of a Holliday junction that undergoes branch migration, cutting at which combination of the sites a to d would generate a crossover? 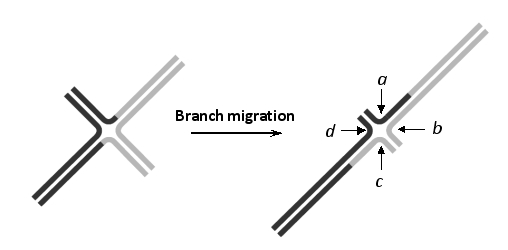
A) a and b
B) a and c
C) a and d
D) b and c
E) b and d

A) a and b
B) a and c
C) a and d
D) b and c
E) b and d

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
22
The sliding clamp and the DNA helicase that function at the replication fork both have three-dimensional structures resembling a ring with a central hole through which DNA is threaded. Which of these proteins, the clamp (C) or the helicase (H), do you think has a wider hole in its structure? Write down C or H as your answer.

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
23
Which of the following is NOT correct regarding homologous recombination and its regulation?
A) Loss of heterozygosity can occur if a broken chromosome is repaired using a sister chromatid instead of its homologous chromosome.
B) Repair of double-strand breaks by homologous recombination is favored during or soon after DNA replication.
C) Homologous recombination can rescue broken or stalled replication forks in S phase.
D) Excessive use of homologous recombination by human cells can lead to cancer.
E) Low usage of homologous recombination by human cells can lead to cancer.
A) Loss of heterozygosity can occur if a broken chromosome is repaired using a sister chromatid instead of its homologous chromosome.
B) Repair of double-strand breaks by homologous recombination is favored during or soon after DNA replication.
C) Homologous recombination can rescue broken or stalled replication forks in S phase.
D) Excessive use of homologous recombination by human cells can lead to cancer.
E) Low usage of homologous recombination by human cells can lead to cancer.

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
24
What combination of the following events normally prevents the origins of replication in the yeast Saccharomyces cerevisiae from "firing" more than once during the cell cycle? Your answer is a two-letter string composed of letters A to E only, e.g. AB. Order the letters in your answer alphabetically.
A) The helicase-loading proteins Cdc6 and Cdt1 are only active in S phase.
B) The helicase Mcm1 can be delivered to the origin recognition complex (ORC) only in S phase.
C) The ORC can only become active (by dephosphorylation) in G? phase.
D) The helicase can only become active (by phosphorylation) in S phase.
E) The ORC can bind to the origin only in S phase.
A) The helicase-loading proteins Cdc6 and Cdt1 are only active in S phase.
B) The helicase Mcm1 can be delivered to the origin recognition complex (ORC) only in S phase.
C) The ORC can only become active (by dephosphorylation) in G? phase.
D) The helicase can only become active (by phosphorylation) in S phase.
E) The ORC can bind to the origin only in S phase.

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
25
Phase variation helps protect the bacterium Salmonella typhimurium against the immune system of its host by switching the orientation of a certain promoter. This process …
A) is carried out through a DNA transposition mechanism.
B) is irreversible.
C) can often result in the excision of the promoter from the chromosome altogether.
D) is mediated by enzymes that form transient covalent bonds with the DNA.
A) is carried out through a DNA transposition mechanism.
B) is irreversible.
C) can often result in the excision of the promoter from the chromosome altogether.
D) is mediated by enzymes that form transient covalent bonds with the DNA.

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
26
In meiosis, a crossover in one position is thought to inhibit crossing-over in the neighboring regions. This regulatory mechanism …
A) results in a very uneven distribution of crossover points along each chromosome.
B) ensures that even small chromosomes undergo at least one crossover.
C) controls how the Holliday junctions are resolved.
D) All of the above.
A) results in a very uneven distribution of crossover points along each chromosome.
B) ensures that even small chromosomes undergo at least one crossover.
C) controls how the Holliday junctions are resolved.
D) All of the above.

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
27
This protein folds into a doughnut shape that can encircle DNA. It can load on the DNA only when the DNA is broken in both strands, so that the DNA can thread through the hole in the protein. Which of the following proteins do you think matches this description?
A) PCNA, the sliding clamp for DNA polymerases at the replication forks
B) Ku, the protein that recognizes DNA ends and can initiate nonhomologous end joining
C) MCM, the helicase critical for the initiation and elongation of replication
D) Topoisomerase II, which can create or relax superhelical tension in DNA
E) RecA/Rad51, which carries out strand invasion in homologous recombination
A) PCNA, the sliding clamp for DNA polymerases at the replication forks
B) Ku, the protein that recognizes DNA ends and can initiate nonhomologous end joining
C) MCM, the helicase critical for the initiation and elongation of replication
D) Topoisomerase II, which can create or relax superhelical tension in DNA
E) RecA/Rad51, which carries out strand invasion in homologous recombination

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
28
DNA-only transposons …
A) can be recognized by the presence of short inverted repeats at each end.
B) often encode a transposase that mediates the transposition process.
C) leave double-strand breaks in the donor chromosome.
D) can move by a cut-and-paste mechanism.
E) All of the above.
A) can be recognized by the presence of short inverted repeats at each end.
B) often encode a transposase that mediates the transposition process.
C) leave double-strand breaks in the donor chromosome.
D) can move by a cut-and-paste mechanism.
E) All of the above.

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
29
In contrast to vertebrates, there is very little DNA methylation in the genomes of invertebrates such as Drosophila melanogaster and Caenorhabditis elegans. Indicate whether you expect each of the following statements to be true (T) or false (F) regarding 5?-CG-3? dinucleotide sequences in the genome. Your answer would be a four-letter string composed of letters T and F only, e.g. TTTT.
( ) On average, approximately one dinucleotide out of every 16 in the human genome is a CG dinucleotide.
( ) On average, approximately one dinucleotide out of every 256 in the D. melanogaster genome is a CG dinucleotide.
( ) The proportion of CG dinucleotides in the human genome is more than that of C. elegans.
( ) The proportion of CG dinucleotides in the human genome is more than that expected by chance.
( ) On average, approximately one dinucleotide out of every 16 in the human genome is a CG dinucleotide.
( ) On average, approximately one dinucleotide out of every 256 in the D. melanogaster genome is a CG dinucleotide.
( ) The proportion of CG dinucleotides in the human genome is more than that of C. elegans.
( ) The proportion of CG dinucleotides in the human genome is more than that expected by chance.

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
30
Which of the following is true regarding retroviral-like retrotransposons?
A) They encode both a reverse transcriptase and an RNA polymerase.
B) They have directly repeated long terminal repeats at their two ends when integrated into chromosomal DNA.
C) Their genomic RNA can be translated to produce viral coat proteins.
D) They leave double-strand breaks in the original donor DNA.
E) The Alu element in our genome is an example of retroviral-like retrotransposons.
A) They encode both a reverse transcriptase and an RNA polymerase.
B) They have directly repeated long terminal repeats at their two ends when integrated into chromosomal DNA.
C) Their genomic RNA can be translated to produce viral coat proteins.
D) They leave double-strand breaks in the original donor DNA.
E) The Alu element in our genome is an example of retroviral-like retrotransposons.

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
31
Which of the following spontaneous lesions in DNA occurs most frequently in a mammalian cell?
A) Depurination
B) Cytosine deamination
C) Guanine oxidation
D) Guanine alkylation
E) Depyrimidination
A) Depurination
B) Cytosine deamination
C) Guanine oxidation
D) Guanine alkylation
E) Depyrimidination

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
32
Which of the following is NOT correct regarding long and short interspersed nuclear elements?
A) Each of them encodes a reverse transcriptase.
B) They rely on the cellular transcription machinery to produce their RNA transcript.
C) They use one of the strands in the target DNA as a primer to synthesize their DNA.
D) Together, they make up about 40% of our genome.
E) They can move into new regions of genome that do not have any homology with their DNA.
A) Each of them encodes a reverse transcriptase.
B) They rely on the cellular transcription machinery to produce their RNA transcript.
C) They use one of the strands in the target DNA as a primer to synthesize their DNA.
D) Together, they make up about 40% of our genome.
E) They can move into new regions of genome that do not have any homology with their DNA.

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
33
Indicate true (T) and false (F) statements below regarding the initiation of replication in human cells. Your answer would be a four-letter string composed of letters T and F only, e.g. TFFF.
( ) Tens of thousands of replication origins are used each time a cell in our body replicates its DNA.
( ) Different cells in our body use different sets of replication origins.
( ) Both replication forks in a replication bubble are normally active in replication.
( ) Gene expression and chromatin structure can affect the choice of the origins to use as well as the order in which they are activated.
( ) Tens of thousands of replication origins are used each time a cell in our body replicates its DNA.
( ) Different cells in our body use different sets of replication origins.
( ) Both replication forks in a replication bubble are normally active in replication.
( ) Gene expression and chromatin structure can affect the choice of the origins to use as well as the order in which they are activated.

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
34
A replication fork is shown schematically below. The strand labeled A is called the … strand.
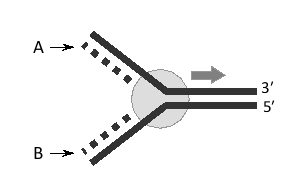


Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
35
What group of mobile genetic elements is largely responsible for the resistance of the modern strains of pathogenic bacteria to common antibiotics?
A) DNA-only transposons
B) Retroviral-like retrotransposons
C) Nonretroviral retrotransposons
D) Site-specific recombinases
A) DNA-only transposons
B) Retroviral-like retrotransposons
C) Nonretroviral retrotransposons
D) Site-specific recombinases

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
36
DNA glycosylases constitute an enzyme family found in all three domains of life. They can …
A) add sugar moieties to DNA.
B) remove sugar moieties from DNA.
C) add a purine or pyrimidine base to DNA.
D) remove a purine or pyrimidine base from DNA.
E) remove a nucleotide from DNA.
A) add sugar moieties to DNA.
B) remove sugar moieties from DNA.
C) add a purine or pyrimidine base to DNA.
D) remove a purine or pyrimidine base from DNA.
E) remove a nucleotide from DNA.

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
37
What are the products of deamination of cytosine and 5-methyl cytosine, respectively?
A) Thymine and uracil
B) Thymine in both cases
C) Uracil and thymine
D) Uracil in both cases
E) Xanthine and hypoxanthine
A) Thymine and uracil
B) Thymine in both cases
C) Uracil and thymine
D) Uracil in both cases
E) Xanthine and hypoxanthine

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
38
In which phases of the eukaryotic cell cycle does homologous recombination often occur to repair DNA damage?
A) G₁ and S phases
B) S and G₂ phases
C) G₂ and M phases
D) M and G₁ phases
E) G₁ and G₂ phases
A) G₁ and S phases
B) S and G₂ phases
C) G₂ and M phases
D) M and G₁ phases
E) G₁ and G₂ phases

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
39
Upon heavy damage to the cell's DNA, the normal replicative DNA polymerases may stall when encountering damaged DNA, triggering the use of backup translesion polymerases. These backup polymerases …
A) lack 3′-to-5′ exonucleolytic proofreading activity.
B) are replaced by the replicative polymerases after adding only a few nucleotides.
C) can create mutations even on undamaged DNA.
D) may recognize specific DNA damage and add the appropriate nucleotide to restore the original sequence.
E) All of the above.
A) lack 3′-to-5′ exonucleolytic proofreading activity.
B) are replaced by the replicative polymerases after adding only a few nucleotides.
C) can create mutations even on undamaged DNA.
D) may recognize specific DNA damage and add the appropriate nucleotide to restore the original sequence.
E) All of the above.

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
40
Which of the following repair pathways can accurately repair a double-strand break?
A) Base excision repair
B) Nucleotide excision repair
C) Direct chemical reversal
D) Homologous recombination
E) Nonhomologous end joining
A) Base excision repair
B) Nucleotide excision repair
C) Direct chemical reversal
D) Homologous recombination
E) Nonhomologous end joining

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
41
Indicate whether each of the following DNA lesions is typically repaired via the base excision (B) or nucleotide excision (N) repair pathway? Your answer would be a four-letter string composed of letters B and N only, e.g. BBBB.
1. Deaminated cytosines
2. Depurinated residues
3. Thymine dimers
4. Bulky guanine adducts
1. Deaminated cytosines
2. Depurinated residues
3. Thymine dimers
4. Bulky guanine adducts

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
42
As shown in the following drawing, a researcher has engineered three pairs of LoxP sites (for conservative site-specific recombination) in a region that contains three reporter genes coding for red, yellow, or cyan fluorescent proteins, respectively. Each type of LoxP sequence (shown as a black, gray, or white arrowhead) is specific, meaning it does not recombine with the other types of LoxP sequences. Upon Cre recombinase activation, depending on which recombination event occurs first (which we assume is random), a number of possible combinations of reporters can remain in the final DNA. For each of the following combinations, indicate whether it can (C) or cannot (N) result from this recombination scheme. Do not consider the re-integration of excised DNA, which happens very rarely. Your answer would be a six-letter string composed of letters C and N only, e.g. CCCCNN.

( ) Red and yellow
( ) Red only
( ) Yellow only
( ) Cyan only
( ) Yellow and cyan
( ) Red and cyan

( ) Red and yellow
( ) Red only
( ) Yellow only
( ) Cyan only
( ) Yellow and cyan
( ) Red and cyan

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
43
Indicate true (T) and false (F) statements below regarding the use of homologous recombination in meiosis. Your answer would be a four-letter string composed of letters T and F only, e.g. FFFF.
( ) Meiotic recombination starts with a double-strand break caused by errors in DNA replication.
( ) Meiotic recombination occurs preferentially between DNA from maternal and paternal chromosome pairs.
( ) Holliday junctions can form during meiotic recombination, sometimes in pairs.
( ) During meiotic recombination in human cells, the majority of the invading strands are released, leading to no crossover.
( ) Meiotic recombination starts with a double-strand break caused by errors in DNA replication.
( ) Meiotic recombination occurs preferentially between DNA from maternal and paternal chromosome pairs.
( ) Holliday junctions can form during meiotic recombination, sometimes in pairs.
( ) During meiotic recombination in human cells, the majority of the invading strands are released, leading to no crossover.

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
44
Indicate true (T) and false (F) descriptions below regarding the nucleotide excision repair pathway. Your answer would be a four-letter string composed of letters T and F only, e.g. FFFF.
( ) It involves recognition of distortions in the DNA double helix rather than specific base changes.
( ) It involves endonucleolytic cleavage and helicase-mediated strand removal.
( ) It involves cleavage by the AP endonuclease.
( ) It is coupled to the DNA transcription machinery of the cell.
( ) It involves recognition of distortions in the DNA double helix rather than specific base changes.
( ) It involves endonucleolytic cleavage and helicase-mediated strand removal.
( ) It involves cleavage by the AP endonuclease.
( ) It is coupled to the DNA transcription machinery of the cell.

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
45
Fill in the blank in the following paragraph. Do NOT use abbreviations.
"In human cells, the predominant pathway to repair double-strand breaks is …, in which the broken ends are simply rejoined with the concomitant loss of a few nucleotides. This leaves scars at the breakage sites. This pathway can potentially create chromosome translocations."
"In human cells, the predominant pathway to repair double-strand breaks is …, in which the broken ends are simply rejoined with the concomitant loss of a few nucleotides. This leaves scars at the breakage sites. This pathway can potentially create chromosome translocations."

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
46
Sort the following steps in the order that they normally happen during the process of repairing double-strand breaks by homologous recombination. Your answer would be a six-letter string composed of letters A to F only, e.g. DEFABC.
(A) Ligation
(B) DNA synthesis using undamaged DNA as the template
(C) DNA synthesis using original DNA as the template
(D) Release of the invading strand
(E) Strand invasion
(F) Nuclease digestion (resection)
(A) Ligation
(B) DNA synthesis using undamaged DNA as the template
(C) DNA synthesis using original DNA as the template
(D) Release of the invading strand
(E) Strand invasion
(F) Nuclease digestion (resection)

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
47
Indicate whether each of the following mobile elements in Drosophila is a DNA-only transposon (D), a retroviral-like retrotransposon (R), or a nonretroviral retrotransposon (N). Your answer would be a four-letter string composed of letters D, R, and N only, e.g. RRRR.
( ) P elements; these have inverted terminal repeats and move with the help of a transposase enzyme.
( ) Copia elements; these have directly repeated long terminal repeats and move with the help of a reverse transcriptase and an integrase.
( ) F elements; these are long interspersed nuclear elements (LINEs).
( ) Mariner elements; these move via a DNA intermediate.
( ) P elements; these have inverted terminal repeats and move with the help of a transposase enzyme.
( ) Copia elements; these have directly repeated long terminal repeats and move with the help of a reverse transcriptase and an integrase.
( ) F elements; these are long interspersed nuclear elements (LINEs).
( ) Mariner elements; these move via a DNA intermediate.

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
48
Consider three types of mobile genetic elements that are found in our genome: the DNA-only transposons (D), the retroviral-like retrotransposons (R), and the nonretroviral retrotransposons (N). Which type appears to still be active and move in our genome, accounting for a detectable fraction of human mutations? Write down your answer as D, R, or N.

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
49
The RecA/Rad51 protein carries out strand exchange during homologous recombination. Indicate true (T) and false (F) statements about this process below. Your answer would be a four-letter string composed of letters T and F only, e.g. FFFF.
( ) RecA hydrolyzes ATP upon binding to the invading DNA strand.
( ) RecA forces the invading strand into a conformation that fully mimics the geometry of a long DNA double helix.
( ) Sampling of the homologous duplex by the invading strand is likely to occur in triplet nucleotide blocks.
( ) RecA-bound, invading, single-stranded DNA binds and destabilizes the homologous duplex to allow the sampling of its sequence by base-pairing.
( ) RecA hydrolyzes ATP upon binding to the invading DNA strand.
( ) RecA forces the invading strand into a conformation that fully mimics the geometry of a long DNA double helix.
( ) Sampling of the homologous duplex by the invading strand is likely to occur in triplet nucleotide blocks.
( ) RecA-bound, invading, single-stranded DNA binds and destabilizes the homologous duplex to allow the sampling of its sequence by base-pairing.

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
50
In cells that are exposed to sunlight, ultraviolet (UV) light can result in covalent linkage between two adjacent DNA bases. If not repaired in time, such dimers can lead to inheritable mutations. Consider the sequence 5?-GGTATGATCATTATAA-3? in the chromosome of a cell that is exposed to intense sunlight. How many possible dimers can form in the region of DNA double helix corresponding to this sequence?

Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck
51
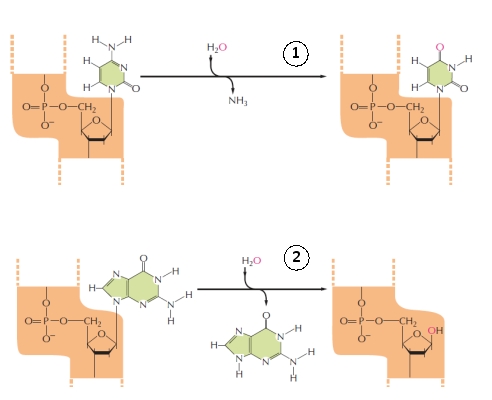
Examples of two general types of DNA damage are shown in the following drawing. Which type of damage (1 or 2) is more common in our cells?


Unlock Deck
Unlock for access to all 51 flashcards in this deck.
Unlock Deck
k this deck


