Deck 7: Control of Gene Expression
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/62
Play
Full screen (f)
Deck 7: Control of Gene Expression
1
Indicate true (T) and false (F) statements below regarding gene control. Your answer would be a four-letter string composed of letters T and F only, e.g. TTFT.
( ) Development of multicellular organisms from a fertilized egg only rarely involves DNA rearrangements in specialized cells.
( ) A typical human cell expresses less than 1% of its approximately 30,000 genes at any given time.
( ) Genes that are expressed in all cell types usually vary in their level of expression in different cell types.
( ) Many differentiated plant cells can be fully de-differentiated and give rise to an entire plant.
( ) Development of multicellular organisms from a fertilized egg only rarely involves DNA rearrangements in specialized cells.
( ) A typical human cell expresses less than 1% of its approximately 30,000 genes at any given time.
( ) Genes that are expressed in all cell types usually vary in their level of expression in different cell types.
( ) Many differentiated plant cells can be fully de-differentiated and give rise to an entire plant.
T
F
T
T
F
T
T
2
The majority of transcription regulators make sequence-specific contacts with DNA in the major groove. In the two diagrams below, where are the contact surfaces that are exposed in the major groove? 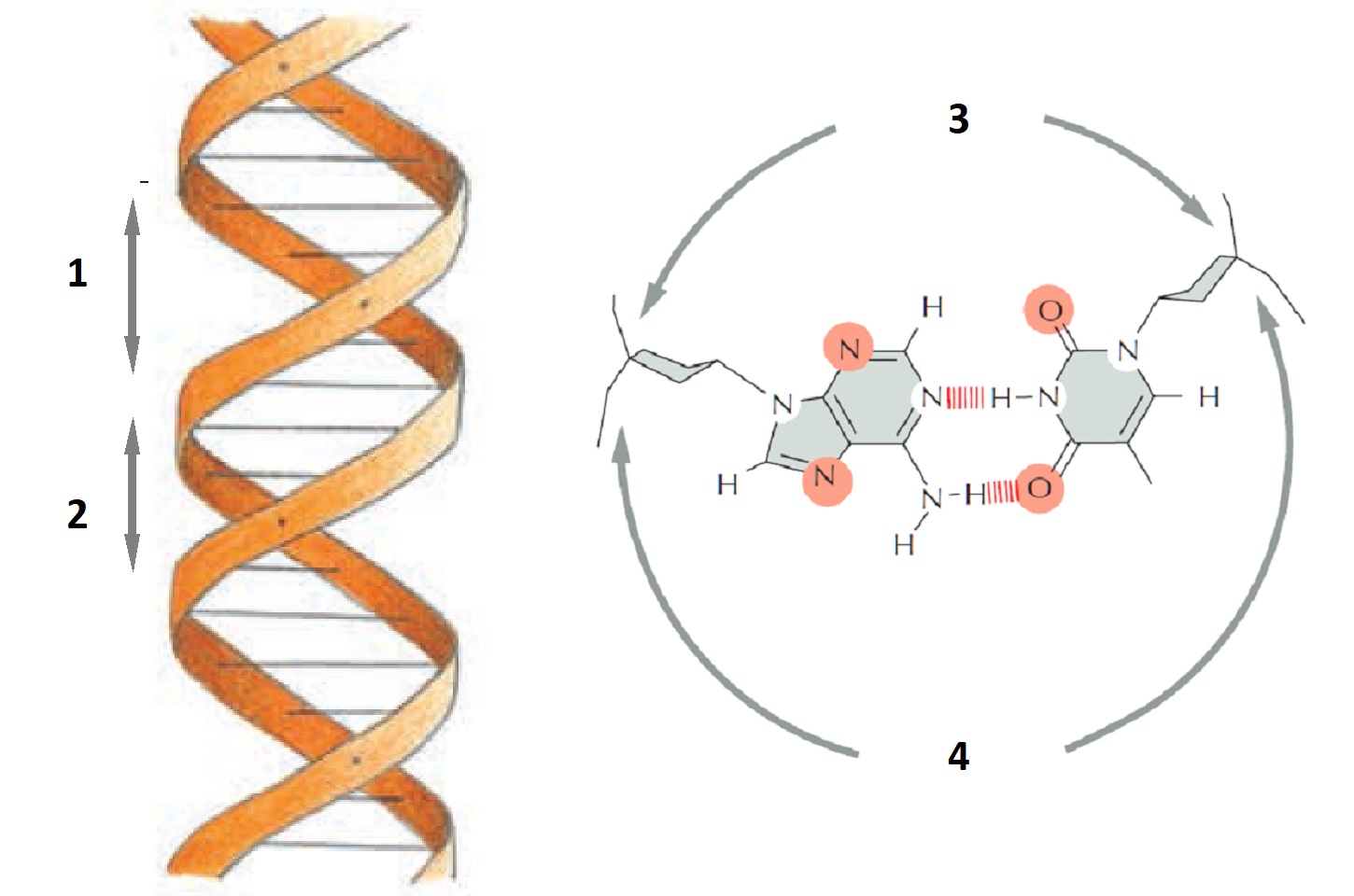
A) 1, 3
B) 1, 4
C) 2, 3
D) 2, 4

A) 1, 3
B) 1, 4
C) 2, 3
D) 2, 4
B
Explanation: Nearly all transcription regulators make the majority of their contacts with the major groove (1 and 4 in the drawings) because the major groove is wider and displays more molecular features than does the minor groove (2 and 3).
Explanation: Nearly all transcription regulators make the majority of their contacts with the major groove (1 and 4 in the drawings) because the major groove is wider and displays more molecular features than does the minor groove (2 and 3).
3
Considering the diagrams below that show hydrogen-bond donors and acceptors as well as hydrophobic groups in four DNA base pairs, which of the following do you think is the most difficult to accomplish by DNA-binding proteins? 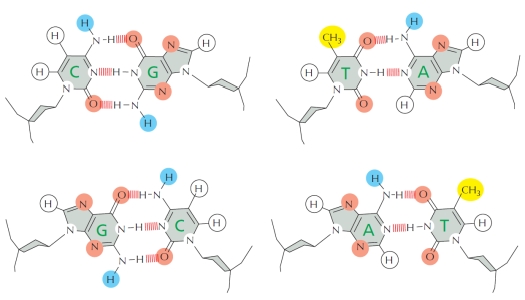
A) Distinguishing between A-T and T-A in the major groove
B) Distinguishing between A-T and C-G in the major groove
C) Distinguishing between C-G and G-C in the major groove
D) Distinguishing between C-G and G-C in the minor groove
E) Distinguishing between C-G and T-A in the minor groove

A) Distinguishing between A-T and T-A in the major groove
B) Distinguishing between A-T and C-G in the major groove
C) Distinguishing between C-G and G-C in the major groove
D) Distinguishing between C-G and G-C in the minor groove
E) Distinguishing between C-G and T-A in the minor groove
D
Explanation: In the minor groove, precise recognition of A-T versus T-A (or C-G versus G-C) is not readily feasible. However, an A-T can still be distinguished from C-G, for example.
Explanation: In the minor groove, precise recognition of A-T versus T-A (or C-G versus G-C) is not readily feasible. However, an A-T can still be distinguished from C-G, for example.
4
Protein subunits that interact specifically with DNA sequences …
A) typically recognize sequences of two to three nucleotide pairs in length.
B) do so via hydrogen bonds, ionic bonds, and hydrophobic interactions.
C) typically form about five weak interactions at the protein-DNA interface.
D) often bind loosely to DNA.
E) All of the above.
A) typically recognize sequences of two to three nucleotide pairs in length.
B) do so via hydrogen bonds, ionic bonds, and hydrophobic interactions.
C) typically form about five weak interactions at the protein-DNA interface.
D) often bind loosely to DNA.
E) All of the above.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
5
Transcription regulation has similarities and differences in bacteria and in eukaryotes. Which of the following is correct in this regard?
A) Most bacterial genes are regulated individually, whereas most eukaryotic genes are regulated in clusters.
B) The rate of transcription for a eukaryotic gene can vary in a much wider range than for a bacterial gene (which is, at most, only about 1000-fold).
C) DNA looping for gene regulation is the rule in bacteria but the exception in eukaryotes.
D) Transcription regulators in both bacteria and eukaryotes usually bind directly to RNA polymerase.
E) The default state of both bacterial and eukaryotic genomes is transcriptionally active.
A) Most bacterial genes are regulated individually, whereas most eukaryotic genes are regulated in clusters.
B) The rate of transcription for a eukaryotic gene can vary in a much wider range than for a bacterial gene (which is, at most, only about 1000-fold).
C) DNA looping for gene regulation is the rule in bacteria but the exception in eukaryotes.
D) Transcription regulators in both bacteria and eukaryotes usually bind directly to RNA polymerase.
E) The default state of both bacterial and eukaryotic genomes is transcriptionally active.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
6
In analysis using two-dimensional gel electrophoresis of the proteins expressed in different cell types, the number of spots that are different in different cells usually exceeds the number of common spots, and even the common spots can still have different intensities. The spots representing which of the following proteins would you expect to be among the common spots when compared across several cell types?
A) RPL10 (a ribosomal protein)
B) HBA1 (a hemoglobin subunit)
C) Insulin (a hormone)
D) Tyrosine aminotransferase (a metabolic enzyme)
E) All of the above
A) RPL10 (a ribosomal protein)
B) HBA1 (a hemoglobin subunit)
C) Insulin (a hormone)
D) Tyrosine aminotransferase (a metabolic enzyme)
E) All of the above

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
7
The following sequence logo represents the preferred cis-regulatory sequences of an imaginary transcription regulator that functions as a dimer. Would you expect this sequence to be recognized by a homodimer (M) or a heterodimer (T)? Write down M or T as your answer.
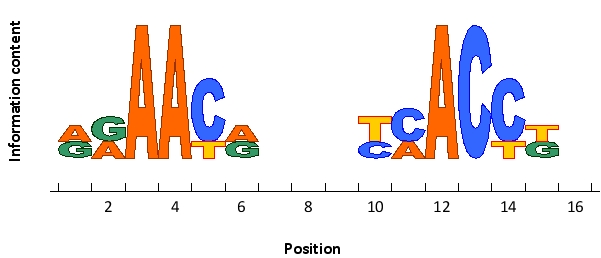


Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
8
Under which of the following conditions is the Lac operon in Escherichia coli fully turned on?
A) Low glucose and lactose levels
B) Low glucose but high lactose levels
C) High glucose but low lactose levels
D) High glucose and lactose levels
E) Low cAMP and high glucose levels
A) Low glucose and lactose levels
B) Low glucose but high lactose levels
C) High glucose but low lactose levels
D) High glucose and lactose levels
E) Low cAMP and high glucose levels

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
9
A DNA-binding protein recognizes a specific eight-nucelotide sequence in DNA. Assuming that its binding is perfectly specific, how many binding sites are expected to exist for it in human genomic DNA, which is composed of about 6 × 10⁹ nucleotide pairs? What about if the protein forms a homodimer? Note that the target sequence can be oriented either way in the double-stranded DNA.
A) About 200,000; about 30,000
B) About 2000; about 300
C) About 200; about 30
D) About 200,000; about 300
E) About 200,000; about 3
A) About 200,000; about 30,000
B) About 2000; about 300
C) About 200; about 30
D) About 200,000; about 300
E) About 200,000; about 3

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
10
Studying the expression of a transcription regulatory protein in two cell types, you have performed experiments showing that the mRNA encoding the protein is present at comparable levels in the cytosol of both cell types. However, based on the expression of its target genes, you suspect that the protein activity might be significantly different in the two cell types. Which of the following steps in expression of the gene encoding this protein is more likely to be differentially controlled in these cell types?
A) Transcription
B) Translation
C) mRNA transport
D) mRNA degradation
A) Transcription
B) Translation
C) mRNA transport
D) mRNA degradation

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
11
Indicate true (T) and false (F) statements below regarding transcription regulators. Your answer would be a four-letter string composed of letters T and F only, e.g. TTTF.
( ) About 10% of the protein-coding genes in most organisms encode transcription regulators.
( ) The packaging of eukaryotic DNA into chromatin dampens the effect of cooperative binding of transcription regulators.
( ) Some DNA-binding proteins can induce bends or kinks in their target DNA.
( ) Dimerization of transcription regulators increases the chance of binding to nonspecific sequences.
( ) About 10% of the protein-coding genes in most organisms encode transcription regulators.
( ) The packaging of eukaryotic DNA into chromatin dampens the effect of cooperative binding of transcription regulators.
( ) Some DNA-binding proteins can induce bends or kinks in their target DNA.
( ) Dimerization of transcription regulators increases the chance of binding to nonspecific sequences.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
12
Which of the following is directed by transcription activators in eukaryotic cells in order to provide a more accessible DNA for the transcription machinery?
A) Nucleosome remodeling
B) Histone removal
C) Histone replacement
D) Histone modifications
E) All of the above
A) Nucleosome remodeling
B) Histone removal
C) Histone replacement
D) Histone modifications
E) All of the above

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
13
Consider a cis-regulatory enhancer sequence in the Escherichia coli chromosome that is located thousands of nucleotide pairs upstream of the gene that it regulates. If the regulatory sequence is mutated to become nonfunctional, the introduction of the wild-type enhancer on a plasmid fails to regulate the gene. This implies that …
A) the regulatory sequence encodes a regulatory protein that binds near the promoter of the target gene and controls RNA polymerase binding.
B) the regulation of the target gene involves looping out of the intervening DNA, and the promoter of the cis-regulatory sequence must be on the same chromosome.
C) the regulatory sequence can bind directly to the RNA polymerase.
D) the regulatory sequence cannot bind to a protein.
A) the regulatory sequence encodes a regulatory protein that binds near the promoter of the target gene and controls RNA polymerase binding.
B) the regulation of the target gene involves looping out of the intervening DNA, and the promoter of the cis-regulatory sequence must be on the same chromosome.
C) the regulatory sequence can bind directly to the RNA polymerase.
D) the regulatory sequence cannot bind to a protein.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
14
The following schematic drawing represents the activation of transcription for a eukaryotic gene (gene X). Indicate what component (A to D) in the drawing corresponds to each of the following. Your answer would be a four-letter string composed of letters A to D only, e.g. ADBC.
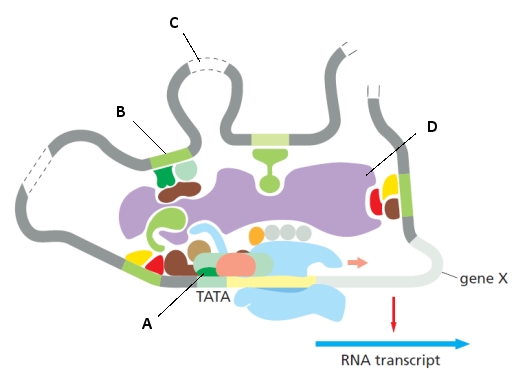
( ) Mediator
( ) cis-Regulatory sequence
( ) A general transcription factor
( ) "Spacer" DNA that may encode lncRNAs

( ) Mediator
( ) cis-Regulatory sequence
( ) A general transcription factor
( ) "Spacer" DNA that may encode lncRNAs

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
15
The Trp operon in Escherichia coli encodes the components necessary for tryptophan biosynthesis. In the presence of the amino acid in a bacterium, …
A) the tryptophan operator is bound to the tryptophan repressor.
B) the tryptophan repressor is bound to bacterial RNA polymerase.
C) the expression of the tryptophan repressor is shut off.
D) the operon genes are expressed.
E) All of the above.
A) the tryptophan operator is bound to the tryptophan repressor.
B) the tryptophan repressor is bound to bacterial RNA polymerase.
C) the expression of the tryptophan repressor is shut off.
D) the operon genes are expressed.
E) All of the above.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
16
In the following schematic graph of a hypothetical set of RNA-seq data, the number of reads is plotted for a region of chromosome containing two genes, from samples obtained from two different tissues. Which gene (X or Y) do you think is more likely a "housekeeping" gene? Which region (1 or 2) within gene Y most likely corresponds to an exon? 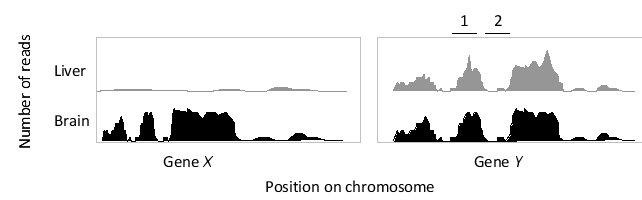
A) Gene X; region 1
B) Gene X; region 2
C) Both genes; region 1
D) Gene Y; region 1
E) Gene Y; region 2

A) Gene X; region 1
B) Gene X; region 2
C) Both genes; region 1
D) Gene Y; region 1
E) Gene Y; region 2

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
17
To prevent spurious transcription from a gene, acetylation of histones-which is carried out by histone acetyl transferases ahead of a moving RNA polymerase II-is quickly reversed by histone deacetylases and histone methyl transferases in the wake of the polymerase, leaving a trail of specific methylated histones. Which of the following curves do you think better represents the distribution of this specific histone methylation mark with respect to a gene? 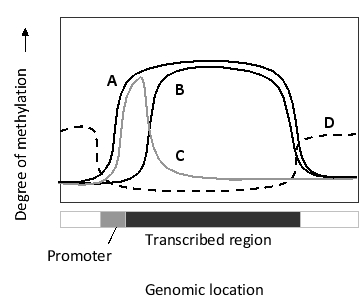


Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
18
A DNA-binding protein binds cooperatively to DNA by fairly weak homodimerization. The binding of this protein to DNA …
A) shows a sigmoidal binding curve.
B) occurs in more of an all-or-none manner, compared to a protein that is a constitutive dimer.
C) occurs despite the fact that the protein molecules are predominantly monomers in solution.
D) All of the above.
A) shows a sigmoidal binding curve.
B) occurs in more of an all-or-none manner, compared to a protein that is a constitutive dimer.
C) occurs despite the fact that the protein molecules are predominantly monomers in solution.
D) All of the above.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
19
What determines the time and place that a certain gene is transcribed in the cell?
A) The type of cis-regulatory sequences associated with it
B) The relative position of cis-regulatory sequences associated with it
C) The arrangement of various cis-regulatory sequences associated with it
D) The specific combination of transcription regulators present in the nucleus
E) All of the above
A) The type of cis-regulatory sequences associated with it
B) The relative position of cis-regulatory sequences associated with it
C) The arrangement of various cis-regulatory sequences associated with it
D) The specific combination of transcription regulators present in the nucleus
E) All of the above

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
20
Which of the following DNA-binding motifs uses β sheets to recognize DNA bases?
A) The helix-turn-helix motif
B) The leucine zipper
C) The zinc finger motif
D) The helix-loop-helix motif
E) None of the above
A) The helix-turn-helix motif
B) The leucine zipper
C) The zinc finger motif
D) The helix-loop-helix motif
E) None of the above

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
21
You have engineered the X chromosomes in female mice such that one X chromosome expresses green fluorescent protein when active, while the other expresses red fluorescent protein. You have used these mice to study cancer in females. You know that each tumor is a clonal cellular proliferation, meaning all of its proliferating cells are descendants of a single original cancer-causing cell. It follows that, unless X-chromosome inactivation is perturbed in tumors, …
A) all tumor cells in one mouse should express the same fluorescent protein (either red or green), but tumor cells from different mice can show either red or green fluorescence.
B) the cells in any tumor should all express the same fluorescent protein (either red or green), but independently derived tumors in the same mouse can show either green or red fluorescence.
C) different cells within each tumor can express different fluorescent proteins, and the tumors would therefore show yellow fluorescence, but each cell shows either red or green fluorescence.
D) each cell can express both fluorescent proteins and would therefore emit yellow fluorescence, and the tumors would glow in yellow as well.
E) different tumors would show red, yellow, or green fluorescence.
A) all tumor cells in one mouse should express the same fluorescent protein (either red or green), but tumor cells from different mice can show either red or green fluorescence.
B) the cells in any tumor should all express the same fluorescent protein (either red or green), but independently derived tumors in the same mouse can show either green or red fluorescence.
C) different cells within each tumor can express different fluorescent proteins, and the tumors would therefore show yellow fluorescence, but each cell shows either red or green fluorescence.
D) each cell can express both fluorescent proteins and would therefore emit yellow fluorescence, and the tumors would glow in yellow as well.
E) different tumors would show red, yellow, or green fluorescence.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
22
Indicate which of the following enzymatic activities corresponds to A, B, or C in the following schematic drawing of DNA methylation events in the human genome. Your answer would be a three-letter string composed of letters A, B, and C only, e.g. CBA.
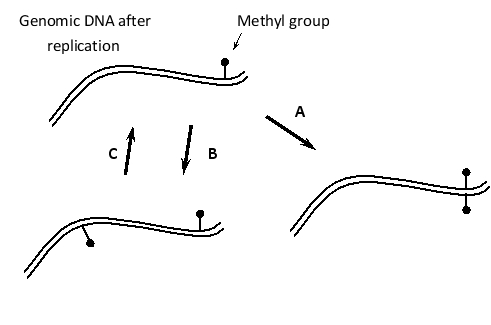
( ) DNA demethylating enzyme
( ) Maintenance methyl transferase enzyme
( ) De novo methyl transferase enzyme

( ) DNA demethylating enzyme
( ) Maintenance methyl transferase enzyme
( ) De novo methyl transferase enzyme

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
23
Indicate true (T) and false (F) statements below regarding DNA methylation in humans. Your answer would be a four-letter string composed of letters T and F only, e.g. TTTF.
( ) Methylation of adenines is the most common DNA methylation in humans.
( ) Methylated cytosine can be accidentally deaminated to produce thymine, leading to a C-to-T transition.
( ) Cytosine methylation often occurs within a 5?-CG-3? sequence.
( ) Shortly after fertilization, a genome-wide wave of demethylation takes place.
( ) Methylation of adenines is the most common DNA methylation in humans.
( ) Methylated cytosine can be accidentally deaminated to produce thymine, leading to a C-to-T transition.
( ) Cytosine methylation often occurs within a 5?-CG-3? sequence.
( ) Shortly after fertilization, a genome-wide wave of demethylation takes place.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
24
Two transcription activators cooperate to recruit a coactivator to a cis-regulatory sequence and activate transcription of a nearby gene. If each of the activators increases the affinity of the coactivator for the reaction site (and therefore the rate of transcription) by 100-fold, how much would you expect the affinity to increase when both activators are bound to DNA compared to when none is bound?
A) 10-fold
B) 100-fold
C) 200-fold
D) 10,000-fold
A) 10-fold
B) 100-fold
C) 200-fold
D) 10,000-fold

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
25
Which of the following is true regarding genomic imprinting?
A) It is an epigenetic phenomenon.
B) It occurs in most animals.
C) It always involves inactivation of genes through direct DNA methylation.
D) It can "unmask" recessive alleles but cannot "mask" dominant ones.
E) All of the above.
A) It is an epigenetic phenomenon.
B) It occurs in most animals.
C) It always involves inactivation of genes through direct DNA methylation.
D) It can "unmask" recessive alleles but cannot "mask" dominant ones.
E) All of the above.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
26
In the following schematic drawings showing four different network motifs in gene transcription circuits, indicate whether each of the motifs 1 to 4 corresponds to a feed-forward loop (E), flip-flop device (F), negative feedback loop (N), or positive feedback loop (P). Your answer would be a four-letter string composed of letters E, F, N, or P only, e.g. PNEF.
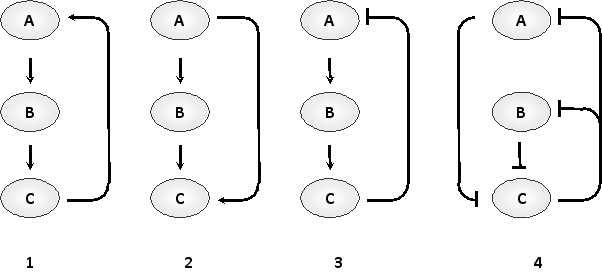


Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
27
Two copies of the gene A exist in a diploid mammal. However, only one copy is expressed in every somatic cell. Different somatic cells in the body of each organism have inactivated one or the other allele in a seemingly random fashion, but when they divide, the daughter cells inherit the choice of the inactive allele faithfully. This is an example of …
A) X-inactivation
B) Genomic imprinting
C) Loss of heterozygosity
D) Monoallelic gene expression
E) Alternative gene splicing
A) X-inactivation
B) Genomic imprinting
C) Loss of heterozygosity
D) Monoallelic gene expression
E) Alternative gene splicing

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
28
Fill in the blank in the following paragraph regarding X-chromosome inactivation. Do not use abbreviations.
"The presence of two copies of the X chromosome in females but only one in males poses a potential problem for regulation of gene expression: everything else being equal, the higher copy number of the X-linked genes can lead to an imbalanced overproduction of their products in females compared to males. Organisms have evolved different … mechanisms to solve this problem. In mammals, for example, the almost complete inactivation of one of the X chromosomes provides the solution to the dosage problem."
"The presence of two copies of the X chromosome in females but only one in males poses a potential problem for regulation of gene expression: everything else being equal, the higher copy number of the X-linked genes can lead to an imbalanced overproduction of their products in females compared to males. Organisms have evolved different … mechanisms to solve this problem. In mammals, for example, the almost complete inactivation of one of the X chromosomes provides the solution to the dosage problem."

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
29
Consider the two feed-forward loops below containing three transcription regulators A, B, and C, where A receives the input signal and C generates the output. In the so-called coherent loop (left), A activates C both directly and indirectly, whereas in an incoherent loop (right), A activates C via one route and inactivates it via the other. Answer the following question(s) based on these network motifs.
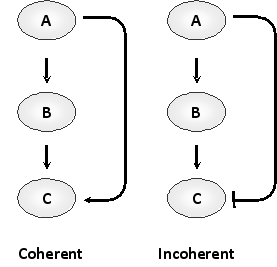
-Would you expect a coherent (C) or an incoherent (I) loop to generate the following response pattern? In this example, A is stimulated by the input, and the transcription of C is measured as the output. Write down C or I as your answer.
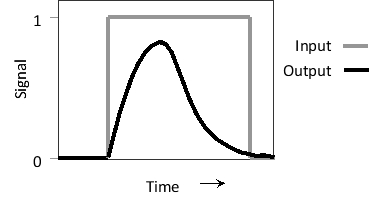

-Would you expect a coherent (C) or an incoherent (I) loop to generate the following response pattern? In this example, A is stimulated by the input, and the transcription of C is measured as the output. Write down C or I as your answer.


Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
30
Expression of the Even-skipped (Eve) gene in early Drosophila embryos is under the control of several transcription regulators. In one example, one of the Eve stripes is positioned near the anterior region of the embryo, and its regulatory module contains binding sites for Bicoid and Hunchback (activators) as well as Giant and Krüppel (inhibitors) such that the gene is expressed only in the region where concentrations of the two activators are high and the concentrations of the two inhibitors are low. A reporter gene can be placed under the control of this module, and it can be shown to form a stripe in the same place in the embryo as the corresponding stripe of Eve. Answer the following question(s) based on these findings.
-In the following schematic diagrams of an early Drosophila embryo, in which region would you expect to find the reporter protein put under the control of the regulatory module mentioned above?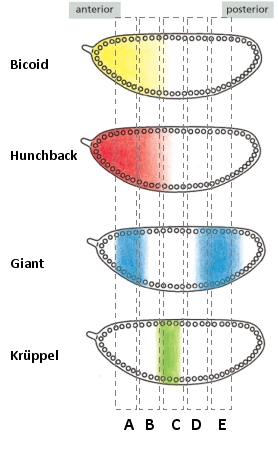
-In the following schematic diagrams of an early Drosophila embryo, in which region would you expect to find the reporter protein put under the control of the regulatory module mentioned above?


Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
31
Consider the two feed-forward loops below containing three transcription regulators A, B, and C, where A receives the input signal and C generates the output. In the so-called coherent loop (left), A activates C both directly and indirectly, whereas in an incoherent loop (right), A activates C via one route and inactivates it via the other. Answer the following question(s) based on these network motifs.

-Considering the two particular feed-forward designs shown, would you expect the coherent (C) or the incoherent (I) loop to have a stable output and respond best to input signals that are above a certain threshold? Write down C or I as your answer.

-Considering the two particular feed-forward designs shown, would you expect the coherent (C) or the incoherent (I) loop to have a stable output and respond best to input signals that are above a certain threshold? Write down C or I as your answer.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
32
Expression of the Even-skipped (Eve) gene in early Drosophila embryos is under the control of several transcription regulators. In one example, one of the Eve stripes is positioned near the anterior region of the embryo, and its regulatory module contains binding sites for Bicoid and Hunchback (activators) as well as Giant and Krüppel (inhibitors) such that the gene is expressed only in the region where concentrations of the two activators are high and the concentrations of the two inhibitors are low. A reporter gene can be placed under the control of this module, and it can be shown to form a stripe in the same place in the embryo as the corresponding stripe of Eve. Answer the following question(s) based on these findings.
-What would you expect to happen to the pattern of reporter expression in flies that lack the gene encoding Bicoid?
A) It would be expressed in all seven stripes.
B) It would be expressed in stripe 5 only.
C) It would expand to cover a broad anterior region of the embryo.
D) It would fail to express efficiently in the stripe 2 region.
E) It would be expressed throughout the whole embryo.
-What would you expect to happen to the pattern of reporter expression in flies that lack the gene encoding Bicoid?
A) It would be expressed in all seven stripes.
B) It would be expressed in stripe 5 only.
C) It would expand to cover a broad anterior region of the embryo.
D) It would fail to express efficiently in the stripe 2 region.
E) It would be expressed throughout the whole embryo.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
33
Expression of the Even-skipped (Eve) gene in early Drosophila embryos is under the control of several transcription regulators. In one example, one of the Eve stripes is positioned near the anterior region of the embryo, and its regulatory module contains binding sites for Bicoid and Hunchback (activators) as well as Giant and Krüppel (inhibitors) such that the gene is expressed only in the region where concentrations of the two activators are high and the concentrations of the two inhibitors are low. A reporter gene can be placed under the control of this module, and it can be shown to form a stripe in the same place in the embryo as the corresponding stripe of Eve. Answer the following question(s) based on these findings.
-In the Eve regulatory modules, the binding sites for Giant and Krüppel are usually close to, or even overlapping with, those of Bicoid and Hunchback. This implies that Eve expression is regulated by … between the activators and regulators.
A) Feedback regulation
B) Cooperation
C) Competition
D) Feed-forward regulation
-In the Eve regulatory modules, the binding sites for Giant and Krüppel are usually close to, or even overlapping with, those of Bicoid and Hunchback. This implies that Eve expression is regulated by … between the activators and regulators.
A) Feedback regulation
B) Cooperation
C) Competition
D) Feed-forward regulation

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
34
Consider the two feed-forward loops below containing three transcription regulators A, B, and C, where A receives the input signal and C generates the output. In the so-called coherent loop (left), A activates C both directly and indirectly, whereas in an incoherent loop (right), A activates C via one route and inactivates it via the other. Answer the following question(s) based on these network motifs.

-Considering coherent and incoherent feed-forward loops, eight different designs are possible using three components and involving activation (positive) and inhibition (negative) regulation only. In all such loops, A would regulate both B and C, and B would regulate only C. How many of these designs constitute a coherent loop? Write down your answer as a number, e.g. 7.

-Considering coherent and incoherent feed-forward loops, eight different designs are possible using three components and involving activation (positive) and inhibition (negative) regulation only. In all such loops, A would regulate both B and C, and B would regulate only C. How many of these designs constitute a coherent loop? Write down your answer as a number, e.g. 7.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
35
Indicate true (T) and false (F) statements below regarding riboswitches. Your answer would be a four-letter string composed of letters T and F only, e.g. TTTF.
( ) A riboswitch permits transcription elongation only when bound to its small-molecule ligand.
( ) Riboswitches appear to be a modern evolutionary invention.
( ) Riboswitches are often located near the 3? end of mRNAs, and therefore fold after the rest of the mRNA.
( ) Riboswitches are exclusively found in prokaryotes.
( ) A riboswitch permits transcription elongation only when bound to its small-molecule ligand.
( ) Riboswitches appear to be a modern evolutionary invention.
( ) Riboswitches are often located near the 3? end of mRNAs, and therefore fold after the rest of the mRNA.
( ) Riboswitches are exclusively found in prokaryotes.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
36
In the following schematic diagram of a region of a mammalian genome, genes A to D (triangles) are located in between a number of insulator elements (white squares) and barrier sequences (black squares). If the cis-regulatory sequence (oval) is bound by an abundant repressor protein, which gene would you expect to be expressed at a higher rate in this cell? 


Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
37
In the following pedigree, females and males are indicated by circles and squares, respectively, and the presence of a rare disease caused by a loss-of-function mutation in an imprinted autosomal gene is indicated by black color. Is the gene maternally imprinted (M; i.e. not expressed from the maternally inherited chromosome) or paternally imprinted (P)? Write down M or P as your answer.
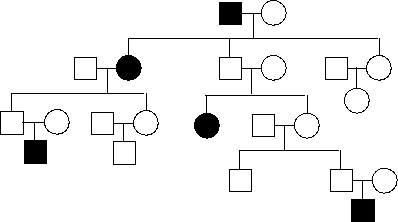


Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
38
Beckwith-Wiedemann syndrome in humans is characterized by "overgrowth" in childhood, sometimes leading to unusually large parts of the body. An imprinted gene cluster on chromosome 11 is associated with this disease. The cluster contains several genes including Igf2 and H19. Igf2 encodes an insulin-like growth factor that is maternally imprinted, i.e. the maternal copy is not expressed. However, the DNA methylation pattern of this locus is not different between the two homologous chromosomes. On the other hand, H19 is also imprinted and its methylation pattern does differ between the two parental chromosomes. H19 is transcribed into a noncoding RNA that appears to silence the transcription of the Igf2 gene in cis. Would you expect the H19 locus to be hypermethylated in the maternally inherited chromosome (M) or paternally inherited chromosome (P)? Write down M or P as your answer.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
39
Expression of the Even-skipped (Eve) gene in early Drosophila embryos is under the control of several transcription regulators. In one example, one of the Eve stripes is positioned near the anterior region of the embryo, and its regulatory module contains binding sites for Bicoid and Hunchback (activators) as well as Giant and Krüppel (inhibitors) such that the gene is expressed only in the region where concentrations of the two activators are high and the concentrations of the two inhibitors are low. A reporter gene can be placed under the control of this module, and it can be shown to form a stripe in the same place in the embryo as the corresponding stripe of Eve. Answer the following question(s) based on these findings.
-What would you expect to happen to the pattern of reporter expression in flies that lack the gene encoding Giant?
A) It would be expressed in all seven stripes.
B) It would be expressed in stripe 5 only.
C) It would expand to cover a broad anterior region of the embryo.
D) It would fail to express efficiently in the stripe 2 region.
E) It would be expressed throughout the whole embryo.
-What would you expect to happen to the pattern of reporter expression in flies that lack the gene encoding Giant?
A) It would be expressed in all seven stripes.
B) It would be expressed in stripe 5 only.
C) It would expand to cover a broad anterior region of the embryo.
D) It would fail to express efficiently in the stripe 2 region.
E) It would be expressed throughout the whole embryo.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
40
Indicate true (T) and false (F) statements below regarding DNA methylation in humans. Your answer would be a four-letter string composed of letters T and F only, e.g. TTTF.
( ) DNA methylation makes repression of gene expression "leakier."
( ) DNA methylation at the promoter region is usually an indication of active transcription.
( ) CG islands are nonpermissive for RNA polymerase assembly.
( ) The dinucleotide 5?-GC-3? is much more common than 5?-CG-3? in the human genome.
( ) DNA methylation makes repression of gene expression "leakier."
( ) DNA methylation at the promoter region is usually an indication of active transcription.
( ) CG islands are nonpermissive for RNA polymerase assembly.
( ) The dinucleotide 5?-GC-3? is much more common than 5?-CG-3? in the human genome.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
41
Which strand in an miRNA precursor will serve as the guide strand in the RNA-induced silencing complex (RISC)? Analysis of miRNA sequences has revealed an asymmetry in the stability of the double-stranded RNA precursor. The strand showing lower thermodynamic stability near its 5? end (nucleotides 2 to 6 in the mature guide strand) is normally selected as the guide strand, and the other strand is usually degraded. In the following miRNA precursor, which strand (1 or 2) do you think will be incorporated into the active RISC? Write down 1 or 2 as your answer.
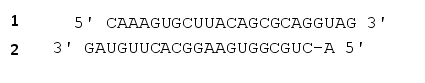
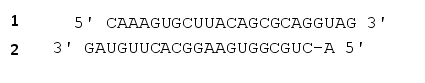

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
42
Indicate whether each of the following descriptions better matches miRNAs (M) or siRNAs (S) in animal cells. Your answer would be a four-letter string composed of letters M and S only, e.g. MMMM.
( ) They usually cleave their target RNAs using the slicing activity of the Argonautes.
( ) They typically show perfect complementarity with their target RNA.
( ) They can bind to the RITS as well as the RISC complex.
( ) They seem to be the more ancient form of small noncoding RNAs.
( ) They usually cleave their target RNAs using the slicing activity of the Argonautes.
( ) They typically show perfect complementarity with their target RNA.
( ) They can bind to the RITS as well as the RISC complex.
( ) They seem to be the more ancient form of small noncoding RNAs.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
43
Stimulated B lymphocytes switch from the synthesis of membrane-bound to secreted antibody molecules by increasing the concentration of a subunit of the trimeric CstF complex that cleaves and polyadenylates mRNAs. How does this up-regulation of CstF bring about the production of soluble antibodies?
A) It favors a strong polyadenylation site in the immunoglobulin primary transcript, creating a longer antibody molecule that is secreted.
B) It activates a weak polyadenylation site in the immunoglobulin primary transcript and prevents splicing, creating a shorter molecule that is secreted.
C) It favors a strong polyadenylation site in the immunoglobulin primary transcript, creating a shorter antibody molecule that is secreted.
D) It activates a weak polyadenylation site in the immunoglobulin primary transcript, creating a longer antibody molecule that is secreted.
E) It favors a strong polyadenylation site in the immunoglobulin primary transcript, aborting translation and creating a shorter antibody molecule that is secreted.
A) It favors a strong polyadenylation site in the immunoglobulin primary transcript, creating a longer antibody molecule that is secreted.
B) It activates a weak polyadenylation site in the immunoglobulin primary transcript and prevents splicing, creating a shorter molecule that is secreted.
C) It favors a strong polyadenylation site in the immunoglobulin primary transcript, creating a shorter antibody molecule that is secreted.
D) It activates a weak polyadenylation site in the immunoglobulin primary transcript, creating a longer antibody molecule that is secreted.
E) It favors a strong polyadenylation site in the immunoglobulin primary transcript, aborting translation and creating a shorter antibody molecule that is secreted.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
44
A certain region of a mammalian genome is transcribed at low but sustained levels using two flanking promoters. The RNA products are annealed together, processed, and used to recruit the RITS complex to this region, as well as chromatin modifying enzymes, RNA-dependent RNA polymerase, and a Dicer enzyme. Indicate whether this pathway is more closely related to that seen in miRNA (M), piRNA (P), or siRNA (S) interference pathways. Write down M, P, or S as your answer.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
45
A researcher studying the human immunodeficiency virus (HIV) has infected human T cells with wild-type or Rev-deficient viruses. She extracts cytoplasmic RNA from the cells and separates each isolated RNA mixture by agarose-gel electrophoresis. She then uses northern analysis using HIV-specific probes to identify those bands that contain viral RNA. The results are presented in the following schematic drawing. Which lane (1 or 2) corresponds to cells infected with the Rev?-deficient virus? What is the size of the full-length HIV genome? 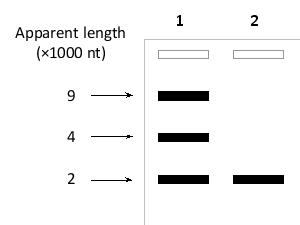
A) Lane 1; about 9000 nucleotides
B) Lane 1; about 4000 nucleotides
C) Lane 1; about 2000 nucleotides
D) Lane 2; about 9000 nucleotides
E) Lane 2; about 2000 nucleotides

A) Lane 1; about 9000 nucleotides
B) Lane 1; about 4000 nucleotides
C) Lane 1; about 2000 nucleotides
D) Lane 2; about 9000 nucleotides
E) Lane 2; about 2000 nucleotides

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
46
What is the function of RNA-dependent RNA polymerases in RNAi?
A) They prevent the spread of the RNAi pathway by replicating the target RNAs.
B) They help amplify the RNAi response by replicating the target RNAs.
C) They produce additional copies of the siRNAs to ensure that the RNAi response is sustained and spread.
D) They are viral proteins that prevent the spread of RNAi by preferentially replicating siRNA sponges.
A) They prevent the spread of the RNAi pathway by replicating the target RNAs.
B) They help amplify the RNAi response by replicating the target RNAs.
C) They produce additional copies of the siRNAs to ensure that the RNAi response is sustained and spread.
D) They are viral proteins that prevent the spread of RNAi by preferentially replicating siRNA sponges.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
47
Which of the following classes of noncoding RNAs is NOT directly involved in RNA interference?
A) miRNA
B) snoRNA
C) piRNA
D) siRNA
A) miRNA
B) snoRNA
C) piRNA
D) siRNA

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
48
In many animals, siRNAs can only function within the cell in which the siRNA is introduced. In contrast, the worm Caenorhabditis elegans can be fed with bacteria that synthesize double-stranded RNAs, and the RNAi spreads throughout the animal. A genetic screen to identify genes involved in systemic RNAi led to the discovery of Sid-1, a transmembrane protein expressed in most tissues in the adult worm. Loss-of-function mutations in the gene encoding Sid-1 restrict the RNAi activity to the cells surrounding the digestive tract after siRNA feeding. Ectopic expression of Sid-1 in Drosophila melanogaster cells that normally lack systemic RNAi and are unable to take up siRNAs from the medium, enables these cells to passively take up siRNA . These findings suggest that …
A) Sid-1 is necessary and sufficient for systemic RNAi and siRNA uptake, respectively.
B) Sid-1 is necessary but not sufficient for systemic RNAi and siRNA uptake, respectively.
C) Sid-1 is not necessary but is sufficient for systemic RNAi and siRNA uptake, respectively.
D) Sid-1 is not necessary and not sufficient for systemic RNAi and siRNA uptake, respectively.
A) Sid-1 is necessary and sufficient for systemic RNAi and siRNA uptake, respectively.
B) Sid-1 is necessary but not sufficient for systemic RNAi and siRNA uptake, respectively.
C) Sid-1 is not necessary but is sufficient for systemic RNAi and siRNA uptake, respectively.
D) Sid-1 is not necessary and not sufficient for systemic RNAi and siRNA uptake, respectively.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
49
The alternative splicing of a certain transcript can result in the production of two mRNA isoforms, one predominant in muscle cells and the other in neurons. The gene contains an exon that is skipped in muscle cells but retained in neurons. You create a mutant version of the gene in which the 3? splice site near this exon is deleted. However, when you introduce this into a culture of neural cells, an even longer pre-mRNA is produced, consistent with the activation of a secondary splice site located near the deleted one. Is the secondary splice site within the exon (E) or its neighboring intron (I)? Write down E or I as your answer.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
50
Indicate true (T) and false (F) statements below regarding P-bodies. Your answer would be a four-letter string composed of letters T and F only, e.g. TTFT.
( ) P-bodies are membrane-enclosed organelles scattered throughout the cytoplasm.
( ) mRNAs that are to be degraded are transferred from P-bodies to stress granules containing decapping and exonuclease enzymes.
( ) When translation initiation is blocked, stress granules typically shrink.
( ) mRNAs associated with stress granules are primed for translation.
( ) P-bodies are membrane-enclosed organelles scattered throughout the cytoplasm.
( ) mRNAs that are to be degraded are transferred from P-bodies to stress granules containing decapping and exonuclease enzymes.
( ) When translation initiation is blocked, stress granules typically shrink.
( ) mRNAs associated with stress granules are primed for translation.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
51
Under iron-starvation conditions in the cell,
A) cytosolic aconitase binds to the 3′ UTR of the mRNA encoding the transferrin receptor.
B) cytosolic aconitase binds to the 3′ UTR of the mRNA encoding ferritin.
C) cytosolic aconitase blocks translation of the mRNA encoding the transferrin receptor.
D) the mRNA encoding the transferrin receptor is cleaved by an endonuclease.
E) All of the above.
A) cytosolic aconitase binds to the 3′ UTR of the mRNA encoding the transferrin receptor.
B) cytosolic aconitase binds to the 3′ UTR of the mRNA encoding ferritin.
C) cytosolic aconitase blocks translation of the mRNA encoding the transferrin receptor.
D) the mRNA encoding the transferrin receptor is cleaved by an endonuclease.
E) All of the above.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
52
When unfolded proteins accumulate in the lumen of the endoplasmic reticulum due to inefficient folding conditions, a membrane-bound protein kinase is activated and phosphorylates a subunit of the translation initiation factor eIF2. Indicate which of the following events would (Y) or would not (N) occur as a result. Your answer would be a four-letter string composed of letters Y and N only, e.g. YYYY.
( ) eIF2 binds tightly to the ribosome.
( ) Translation initiation is stimulated.
( ) The GDP-bound form of eIF2 accumulates.
( ) eIF2B is activated.
( ) eIF2 binds tightly to the ribosome.
( ) Translation initiation is stimulated.
( ) The GDP-bound form of eIF2 accumulates.
( ) eIF2B is activated.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
53
Some viruses encode a protease that cleaves the translation initiation factor eIF4G, rendering it unable to bind to eIF4E. What is the consequence of this cleavage?
A) It shuts down most of the cellular translation machinery, which causes the release of virus particles.
B) It favors viral protein synthesis because IRES-dependent translation initiation is inhibited.
C) It shuts down most host-cell protein synthesis and diverts the translation machinery to IRES-dependent initiation, thus favoring viral protein synthesis.
D) It favors viral protein synthesis by shutting down translation from uORFs.
E) It shuts down IRES-dependent translation, forcing the virus into latency.
A) It shuts down most of the cellular translation machinery, which causes the release of virus particles.
B) It favors viral protein synthesis because IRES-dependent translation initiation is inhibited.
C) It shuts down most host-cell protein synthesis and diverts the translation machinery to IRES-dependent initiation, thus favoring viral protein synthesis.
D) It favors viral protein synthesis by shutting down translation from uORFs.
E) It shuts down IRES-dependent translation, forcing the virus into latency.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
54
The schematic diagram below shows the processing of a class of small RNAs that are involved in RNA interference (RNAi). Which of the following is true regarding these RNAs? 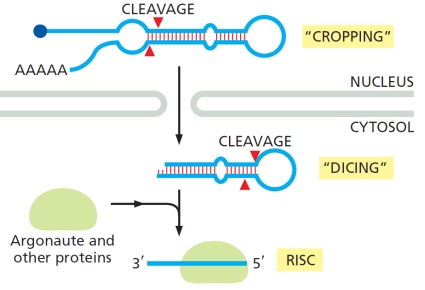


Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
55
Indicate true (T) and false (F) statements below regarding RNA editing. Your answer would be a four-letter string composed of letters T and F only, e.g. TTTF.
( ) RNA editing in animal cells mainly takes place through adenine or cytosine deamination.
( ) RNA editing can change the pattern of pre-mRNA splicing.
( ) RNA editing in coding regions can result in changes in the protein sequence.
( ) Retroviruses such as HIV encode RNA-editing components to confront the host defense mechanisms.
( ) RNA editing in animal cells mainly takes place through adenine or cytosine deamination.
( ) RNA editing can change the pattern of pre-mRNA splicing.
( ) RNA editing in coding regions can result in changes in the protein sequence.
( ) Retroviruses such as HIV encode RNA-editing components to confront the host defense mechanisms.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
56
You have transfected HeLa cells with a gene encoding the human immunodeficiency virus (HIV) protein Rev and have induced the expression of the protein. You incubate the cell culture in the presence or absence of leptomycin B and later measure the localization of Rev inside the cells by immunofluorescence microscopy. Leptomycin B specifically inhibits the cellular Crm1 protein, a nuclear transport receptor that is essential for the normal function of Rev in the HIV life cycle. Your results are tabulated below. Which condition (1 or 2) do you think corresponds to the presence of leptomycin B?
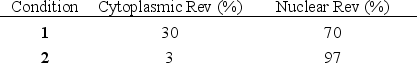
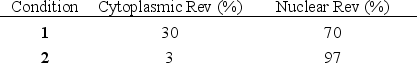

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
57
Imagine a gene encoding a pre-mRNA with twelve exons, ten of which can be alternatively spliced in vivo. Assuming that alternative splicing for this RNA only occurs through exon skipping, how many different proteins can possibly be made from this pre-mRNA as a result of alternative splicing?
A) About 10
B) About 20
C) About 100
D) About 200
E) About 1000
A) About 10
B) About 20
C) About 100
D) About 200
E) About 1000

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
58
In the following schematic diagram of five pairs of alternative RNA splicing patterns, which pair depicts exon skipping only? In exon skipping, an exon is spliced out of the RNA transcript along with its flanking introns. In the diagram, exons are colored dark blue and introns are colored yellow. Light blue indicates possible exons. 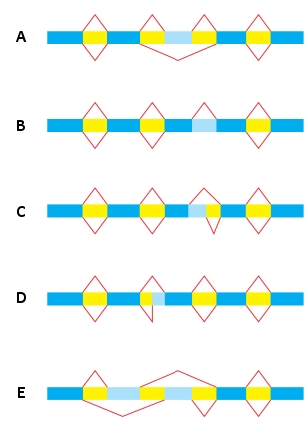


Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
59
Indicate true (T) and false (F) statements below regarding mRNA stability and degradation. Your answer would be a four-letter string composed of letters T and F only, e.g. TTFT.
( ) Any factor that affects the translation efficiency of an mRNA tends to have the opposite effect on its degradation.
( ) Decapping of mRNA normally occurs after poly-A tail shortening removes the poly-A tail and starts chewing into the 3?-UTR.
( ) Most mRNA decay in the cells generally proceeds via endonucleolytic cleavage.
( ) As a general rule, eukaryotic mRNAs have a shorter half-life compared to bacterial mRNAs.
( ) Any factor that affects the translation efficiency of an mRNA tends to have the opposite effect on its degradation.
( ) Decapping of mRNA normally occurs after poly-A tail shortening removes the poly-A tail and starts chewing into the 3?-UTR.
( ) Most mRNA decay in the cells generally proceeds via endonucleolytic cleavage.
( ) As a general rule, eukaryotic mRNAs have a shorter half-life compared to bacterial mRNAs.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
60
Gcn4 is a protein kinase that can phosphorylate and inactivate the initiation factor eIF2. The Gcn4 mRNA contains several short upstream open reading frames (uORFs) that negatively regulate its translation initiation. The phosphorylation of eIF2 by Gcn4, therefore, can control Gcn4 expression …
A) in a negative feedback loop.
B) in a positive feedback loop.
C) in a feed-forward loop.
A) in a negative feedback loop.
B) in a positive feedback loop.
C) in a feed-forward loop.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
61
Consider the descriptions below for three different lncRNAs, and determine whether they function in cis (C) or in trans (T). Your answer would be a three-letter string composed of letters C and T only, e.g. CCT.
( ) The tsiX RNA is transcribed from the X-inactivation center in the X chromosome that will not be inactivated. It is antisense with respect to Xist, repressing its transcription and function.
( ) The ANRIL lncRNA is an antisense transcript in a gene cluster expressing inhibitors of cell-cycle proteins. It silences the entire cluster by recruiting heterochromatin proteins of the Polycomb group.
( ) The PTENP1 lncRNA is similar to an mRNA that encodes the protein PTEN, and contains many of the miRNA response elements present in PTEN mRNA; it therefore protects the PTEN mRNA from miRNA-mediated silencing by acting as a decoy.
( ) The tsiX RNA is transcribed from the X-inactivation center in the X chromosome that will not be inactivated. It is antisense with respect to Xist, repressing its transcription and function.
( ) The ANRIL lncRNA is an antisense transcript in a gene cluster expressing inhibitors of cell-cycle proteins. It silences the entire cluster by recruiting heterochromatin proteins of the Polycomb group.
( ) The PTENP1 lncRNA is similar to an mRNA that encodes the protein PTEN, and contains many of the miRNA response elements present in PTEN mRNA; it therefore protects the PTEN mRNA from miRNA-mediated silencing by acting as a decoy.

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck
62
Comparing the bacterial CRISPR system and the eukaryotic RNAi mediated by siRNAs, indicate whether each of the following CRISPR components is most analogous to Argonaute (A), siRNA (S), or target mRNA (T) in the RNAi pathway. Your answer would be a three-letter string composed of letter A, S, and T only.
( ) Cas
( ) Viral DNA
( ) crRNA
( ) Cas
( ) Viral DNA
( ) crRNA

Unlock Deck
Unlock for access to all 62 flashcards in this deck.
Unlock Deck
k this deck


