Deck 8: Analyzing Cells, Molecules, and Systems
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/95
Play
Full screen (f)
Deck 8: Analyzing Cells, Molecules, and Systems
1
For the two following graphs, the same purified protein has been subjected to velocity sedimentation and gel filtration, as indicated. Two absorption peaks are visible in each graph, corresponding to the monomeric and dimeric forms of the protein. Which peaks correspond to the protein dimer? 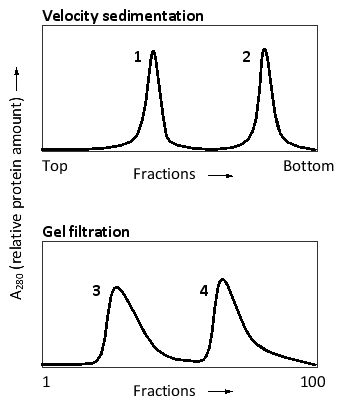
A) 1 and 3
B) 1 and 4
C) 2 and 3
D) 2 and 4

A) 1 and 3
B) 1 and 4
C) 2 and 3
D) 2 and 4
C
Explanation: The larger dimer would sediment faster and would appear near the bottom of the ultracentrifuge tube (peak 2). In gel filtration, the dimer would come off the column in earlier fractions (peak 3).
Explanation: The larger dimer would sediment faster and would appear near the bottom of the ultracentrifuge tube (peak 2). In gel filtration, the dimer would come off the column in earlier fractions (peak 3).
2
Three subunits of a protein complex have been resolved in a two-dimensional polyacrylamide gel, as shown in the schematic drawing below. Indicate whether each of the following better describes the a, b, or c subunit. Your answer would be a three-letter string composed of letters a, b, and c only, e.g. cca.
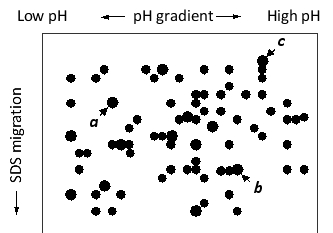
( ) It has the smallest apparent molecular weight.
( ) It has the highest negative charge.
( ) It has the lowest isoelectric point.

( ) It has the smallest apparent molecular weight.
( ) It has the highest negative charge.
( ) It has the lowest isoelectric point.
b
a
a
a
a
3
Indicate whether each of the following descriptions applies to transformed cell lines (T), nontransformed cell lines (N), or both (B). Your answer would be a four-letter string composed of letters T, N, and B only, e.g. BBNN.
( ) They can be thawed and recultured after long-term storage in liquid nitrogen.
( ) They often grow only when attached to a surface, such as that of a culture dish.
( ) They proliferate to very high densities.
( ) They usually cause tumors when injected into immunocompromised animals.
( ) They can be thawed and recultured after long-term storage in liquid nitrogen.
( ) They often grow only when attached to a surface, such as that of a culture dish.
( ) They proliferate to very high densities.
( ) They usually cause tumors when injected into immunocompromised animals.
B
N
T
T
N
T
T
4
What are the advantages of monoclonal antibodies over antisera?
A) They can be produced in higher quantities.
B) They can be produced with higher purity.
C) They have a more uniform specificity.
D) They can be made against molecules that constitute only a minor component of a complex mixture.
E) All of the above.
A) They can be produced in higher quantities.
B) They can be produced with higher purity.
C) They have a more uniform specificity.
D) They can be made against molecules that constitute only a minor component of a complex mixture.
E) All of the above.

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
5
In SDS-PAGE of proteins, …
A) the proteins are unfolded while being separated.
B) the proteins are separated by size, mostly independent of their native charge.
C) large proteins move more slowly.
D) an ionic detergent is used.
E) All of the above.
A) the proteins are unfolded while being separated.
B) the proteins are separated by size, mostly independent of their native charge.
C) large proteins move more slowly.
D) an ionic detergent is used.
E) All of the above.

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
6
In isoelectric focusing, is the low-pH end close to the anode (A; positive electrode) or cathode (C)? Would you use octylglucoside (O; a nonionic detergent) or SDS (S) for separation of proteins by this technique? Would a highly basic protein be near the high-pH (H) or low-pH (L) end of the strip at the end of the experiment? Write down your answer as a three-letter string using the letters in the parentheses above, e.g. ASH.

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
7
Which of the following chromatography methods resembles immunoprecipitation in the purification of proteins from mixtures?
A) Ion-exchange chromatography
B) Gel-filtration chromatography
C) Hydrophobic interaction chromatography
D) Affinity chromatography
A) Ion-exchange chromatography
B) Gel-filtration chromatography
C) Hydrophobic interaction chromatography
D) Affinity chromatography

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
8
Indicate whether each of the following descriptions better applies to equilibrium sedimentation (E) or velocity sedimentation (V). Your answer would be a four-letter string composed of letters E and V only, e.g. EEEE.
( ) It separates subcellular components based on their buoyant density.
( ) It can conveniently separate 15N-containing and 14N-containing DNA.
( ) It uses a shallow sucrose gradient to stabilize the sedimentation bands against convection.
( ) In this method, subcellular components may either move up or down to form discrete bands.
( ) It separates subcellular components based on their buoyant density.
( ) It can conveniently separate 15N-containing and 14N-containing DNA.
( ) It uses a shallow sucrose gradient to stabilize the sedimentation bands against convection.
( ) In this method, subcellular components may either move up or down to form discrete bands.

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
9
Fill in the blank in the following paragraph regarding plant cell cultures. Do not use abbreviations.
"Plant cells can be easily rendered totipotent to give rise to all plant tissues on their own. This can be done by culturing a plant tissue in sterile nutrient media to produce an amorphous cell mass called a …"
"Plant cells can be easily rendered totipotent to give rise to all plant tissues on their own. This can be done by culturing a plant tissue in sterile nutrient media to produce an amorphous cell mass called a …"

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
10
You have used fluorescence-activated cell sorting to separate blood T lymphocytes from a healthy person and a patient infected with a virus. To sort the cells, you used antibodies against two cell-surface receptors, CD4 and CD8, each conjugated to a different fluorescent dye. Based on the results below, which cell type do you think is depleted by the viral infection: CD4+ helper T cells (H) or CD8+ cytotoxic T cells (C)? In these dot plots, each cell is represented by a dot whose coordinates are the intensities of CD4 and CD8 signals. Write down H or C as your answer.
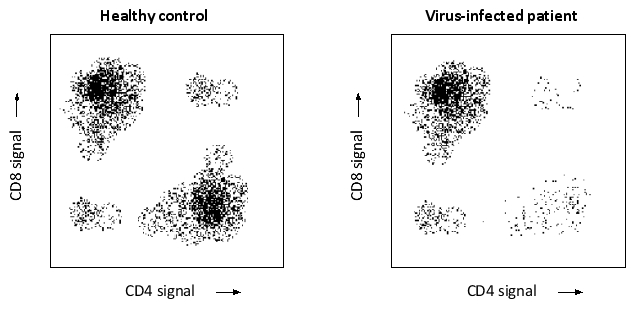


Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
11
Migration of proteins during separation by SDS polyacrylamide-gel electrophoresis (SDS-PAGE) is generally related to the logarithm of their molecular weight, as depicted in the qualitative graph below. If a protein mixture containing proteins of evenly distributed molecular weights (e.g. 10, 20, 30, 40, etc.) is separated by SDS-PAGE, which of the following staining patterns would you expect to observe? 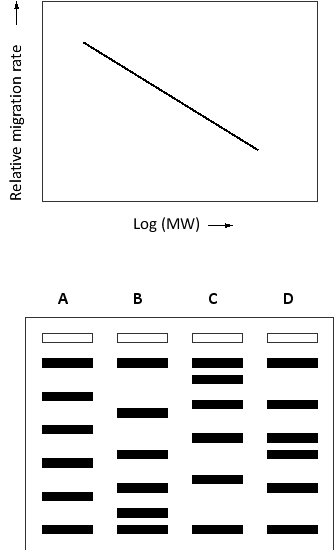


Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
12
In purifying proteins by column chromatography, elution can be performed in different ways, depending on the type of matrix. For example, bound proteins can be eluted by gradually increasing or decreasing salt concentration in the solution applied to the column. For which of the following matrices do you think increasing and decreasing salt concentration, respectively, are used for elution?
A) Ion-exchange; hydrophobic
B) Affinity; ion-exchange
C) Hydrophobic; affinity
D) Hydrophobic; ion-exchange
E) Ion-exchange; gel-filtration
A) Ion-exchange; hydrophobic
B) Affinity; ion-exchange
C) Hydrophobic; affinity
D) Hydrophobic; ion-exchange
E) Ion-exchange; gel-filtration

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
13
Sort the following cellular components to reflect the centrifugal force required to sediment them by preparative ultracentrifugation, from low to high force required. Your answer would be a four-letter string composed of letters R, M, L, and N, e.g. LRNM.
(L) Lysosomes
(M) Microsomes
(N) Nuclei
(R) Ribosomes (free cytosolic)
(L) Lysosomes
(M) Microsomes
(N) Nuclei
(R) Ribosomes (free cytosolic)

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
14
Where do individual peptides get fragmented in a tandem mass spectrometry (MS/MS) set-up?
A) Electrospray ion source
B) Mass filter
C) Inert gas chamber
D) Mass analyzer
E) Detector
A) Electrospray ion source
B) Mass filter
C) Inert gas chamber
D) Mass analyzer
E) Detector

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
15
You have discovered a mutant protein that forms promiscuous disulfide bonds with many other cellular proteins in the endoplasmic reticulum. Using an antibody against the protein, you immunoprecipitate proteins that interact with it. You then separate these proteins using a special type of two-dimensional polyacrylamide-gel electrophoresis (PAGE): in this method, separation in the first dimension is carried out by SDS-PAGE in the absence of β-mercaptoethanol, while separation in the second dimension is carried out by SDS-PAGE in the presence of β-mercaptoethanol. The result is shown in the following schematic drawing. Apparent molecular weights are indicated on the left. What do you think is the molecular weight of the mutant protein? What is the molecular weight represented by the question mark? 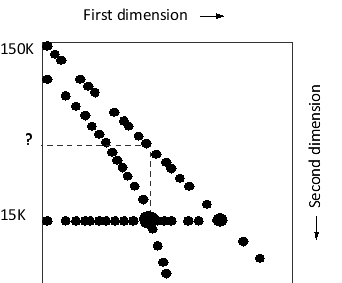
A) 15K, 45K
B) 30K, 45K
C) 15K, 30K
D) 30K, 30K
E) 30K, 60K

A) 15K, 45K
B) 30K, 45K
C) 15K, 30K
D) 30K, 30K
E) 30K, 60K

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
16
How is tandem affinity purification distinct from other purification schemes such as simple affinity or ion-exchange chromatography?
A) In its use of antibodies
B) In its use of proteases
C) In its use of glutathione S-transferase
D) In its use of viral peptides
E) In its use of limited washing steps
A) In its use of antibodies
B) In its use of proteases
C) In its use of glutathione S-transferase
D) In its use of viral peptides
E) In its use of limited washing steps

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
17
How are antibody-producing hybridoma cell lines immortalized to provide an unlimited source of monoclonal antibodies?
A) By ectopic expression of telomerase
B) By transformation with a retrovirus
C) By cell fusion
D) By overproduction of introduced oncogenes
E) By irradiation
A) By ectopic expression of telomerase
B) By transformation with a retrovirus
C) By cell fusion
D) By overproduction of introduced oncogenes
E) By irradiation

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
18
Which mass spectrometry instrument is most suitable for determining all the proteins present in an organelle?
A) MALDI-TOF
B) MS/MS
C) LC-MS/MS
D) GC-MS
A) MALDI-TOF
B) MS/MS
C) LC-MS/MS
D) GC-MS

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
19
Due to friction with molecules of the surrounding solution, sedimenting particles in a mixture reach a terminal velocity when subjected to ultracentrifugation. The sedimentation coefficient (S) for a particle is defined as the ratio of terminal velocity (vt) to the centrifugal acceleration applied to the particle (ac). S = vt / ac
If each Svedberg unit (S value) is defined as 10-¹³ seconds, approximately how long would it take for a free 50S ribosomal subunit to travel the length of a 9 cm ultracentrifuge tube subjected to a centrifugal force of 50,000 times gravity (g)? Recall that g is approximately 10 m/s².
A) 1 min
B) 10 min
C) 50 min
D) 5 h
E) 10 h
If each Svedberg unit (S value) is defined as 10-¹³ seconds, approximately how long would it take for a free 50S ribosomal subunit to travel the length of a 9 cm ultracentrifuge tube subjected to a centrifugal force of 50,000 times gravity (g)? Recall that g is approximately 10 m/s².
A) 1 min
B) 10 min
C) 50 min
D) 5 h
E) 10 h

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
20
To create cellular factories for monoclonal antibody production, cells derived from an immortalized B lymphocyte cell line (M) are cultured together with antibody-secreting B lymphocytes (B) on a so-called HAT medium. The medium contains an inhibitor of DHFR, an enzyme essential for de novo nucleotide biosynthesis. Additionally, it contains substrates for the only alternative nucleotide biosynthetic pathway called the salvage pathway. The enzyme HGPRT is essential for the salvage pathway. One of the cells mentioned above lacks the gene encoding one of the enzymes mentioned. Considering the role of the HAT medium in selecting for hybrid cells that are both immortalized and secrete antibodies, which cell (B or M) do you think is missing an enzyme? Which enzyme is missing in this cell?
A) M - DHFR
B) M - HGPRT
C) B - DHFR
D) B - HGPRT
A) M - DHFR
B) M - HGPRT
C) B - DHFR
D) B - HGPRT

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
21
You have devised genetic screens to identify gain- and loss-of-function mutations in the Escherichia coli DNA ligase gene. One of the screens (1) seeks mutant cells that are infected more readily with a ligase-deficient strain of T4 phage. The other screen (2) seeks mutant cells that have increased radiation sensitivity at high temperatures. Indicate whether each of the following descriptions better applies to screen 1 (1) or screen 2 (2). Your answer would be a three-digit number composed of digits 1 and 2 only, e.g. 122.
( ) It would identify gain-of-function DNA ligase mutants.
( ) It would identify loss-of-function DNA ligase mutants.
( ) It would identify conditional mutants.
( ) It would identify gain-of-function DNA ligase mutants.
( ) It would identify loss-of-function DNA ligase mutants.
( ) It would identify conditional mutants.

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
22
In a forensic lab, DNA samples from a mother (M) and her child (C) have been analyzed at three short tandem repeat (STR) loci using PCR and gel electrophoresis. The same analysis has been performed for five men (1 to 5). According to the results below, which men can be eliminated as possible candidates for the biological father of the child? Your answer would be a number composed of digits 1 to 5 only, e.g. 145.
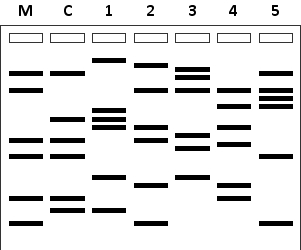


Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
23
The following schematic graph shows temperature change over time in a cycle of a PCR assay. Which steps (A to C) in the cycle correspond to hybridization (annealing), strand separation (denaturation), and DNA synthesis (extension), respectively? Your answer would be a three-letter string composed of letters A to C, e.g. ACB.
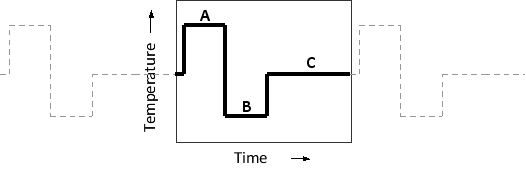


Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
24
In an unfolded (random coil) protein, amino acid residues are exposed to the solvent and share more or less the same environment, whereas each residue in a folded protein has its own unique neighborhood. This fact can be exploited in using NMR to study protein folding. The two schematic diagrams below represent two-dimensional NMR spectra for the same protein in either its folded (native) or unfolded state. The chemical shifts, which depend on the local neighborhood of each atom, are plotted in these diagrams. Which diagram (A or B) do you think corresponds to the folded protein? Write down A or B as your answer. 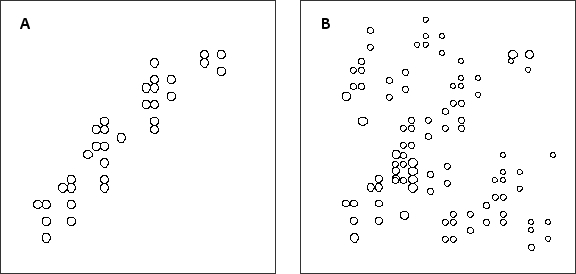


Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
25
Indicate whether each of the following manipulations or procedures used in current recombinant DNA technology commonly rely on an enzyme (E) or are done nonenzymatically (N). Your answer would be a six-letter string composed of letters E and N only, e.g. EEEEEE.
( ) Cleavage of DNA at specific sites
( ) DNA ligation
( ) DNA cloning
( ) Nucleic acid hybridization
( ) Synthesis of DNA of any desired specific sequence
( ) DNA sequencing
( ) Cleavage of DNA at specific sites
( ) DNA ligation
( ) DNA cloning
( ) Nucleic acid hybridization
( ) Synthesis of DNA of any desired specific sequence
( ) DNA sequencing

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
26
Your friend has cut a 3000-nucleotide-pair circular plasmid with either or both restriction enzymes A and B, and has separated the digestion products by electrophoresis. The results are shown in the schematic drawing below, with the approximate size of the fragments indicated above each band. Based on these results, what is the closest distance (in nucleotide pairs) between an A restriction site and a B restriction site on this plasmid? 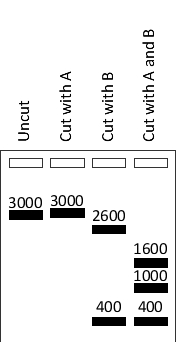
A) 400
B) 600
C) 1000
D) 1200
E) 1600

A) 400
B) 600
C) 1000
D) 1200
E) 1600

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
27
You have carried out a genetic screen in a species of fish, and have identified a new gene in this organism. You have cloned and sequenced the gene, but do not know anything about the biochemical function of the gene product. Which of the following is the most sensible next step in order to get clues about the function?
A) Purifying the protein product and determining its structure by x-ray diffraction
B) Screening for small molecules that perturb gene function
C) Using the BLAST algorithm to scan sequence databases for similar sequences
D) Using MS/MS to sequence the gene product without the need for purification
E) Mutating the gene and performing reverse genetics
A) Purifying the protein product and determining its structure by x-ray diffraction
B) Screening for small molecules that perturb gene function
C) Using the BLAST algorithm to scan sequence databases for similar sequences
D) Using MS/MS to sequence the gene product without the need for purification
E) Mutating the gene and performing reverse genetics

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
28
You have used a variation of fluorescence in situ hybridization (FISH) called flow-FISH. In this technique, DNA probes are labeled with fluorescence and hybridized to their targets inside the cells in a tissue. The cells are then sorted using a fluorescence-activated cell sorter based on their fluorescent signal intensity. The probes in your experiment are designed to hybridize to human telomeric sequences at the ends of the chromosomes. You perform flow-FISH using these probes on two human cell types, an early primary fibroblast culture and a late secondary culture that is showing the signs of replicative cell senescence. The sorting results are presented in the following histograms. Which histogram (1 or 2) would you expect to correspond to the primary culture?
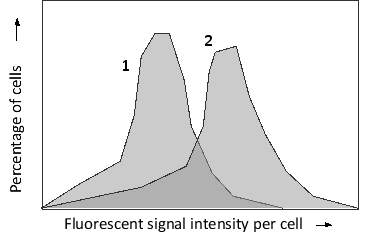


Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
29
The specificity of nucleic acid hybridization is remarkable. Even a single mismatch between a probe and a target sequence can be detected depending on hybridization conditions. This sensitivity can be employed in applications such as SNP haplotyping. Consider a probe that is complementary to a 20-nucleotide-long genomic region including a single polymorphic nucleotide. The probe is perfectly complementary to the more common allele, but has one mismatch in the case of the minor allele. The temperature-dependent hybridization of the probe to the single-strand genomic fragment is schematically presented in the following graph. To detect hybridization, a fluorescent dye is added to the reaction. The dye fluoresces strongly only when intercalated between the bases of double-stranded DNA, and is therefore used to quantify double-strand formation or dissociation. Which "melting curve" (1 or 2) in the graph do you think corresponds to the more common SNP allele? Which curve shows a higher melting temperature? 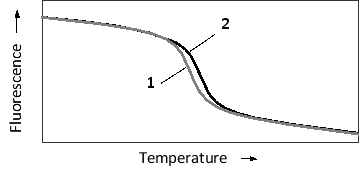
A) 1; 1
B) 1; 2
C) 2; 1
D) 2; 2

A) 1; 1
B) 1; 2
C) 2; 1
D) 2; 2

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
30
The binding of protein A to two other proteins, B and C, has been studied with fluorescence anisotropy, and the results of the measurements have been plotted in the following schematic diagram. Given equal kₒn rate constants, which complex do you think has a longer half-life? What is the value of K?ₐ (the association constant) for this complex? 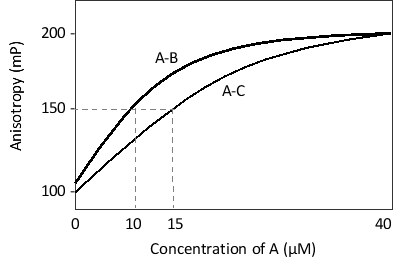
A) A-B; 10⁵ L/mol
B) A-B; 10-⁵ L/mol
C) A-C; 1.5 × 10⁵ L/mol
D) A-C; 6.7 × 10⁴ L/mol
E) A-C; 6.7 × 10-⁴ L/mol

A) A-B; 10⁵ L/mol
B) A-B; 10-⁵ L/mol
C) A-C; 1.5 × 10⁵ L/mol
D) A-C; 6.7 × 10⁴ L/mol
E) A-C; 6.7 × 10-⁴ L/mol

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
31
Indicate whether each of the following descriptions better applies to the determination of atomic structure of proteins by x-ray diffraction (X) or by nuclear magnetic resonance (NMR) spectroscopy (N). Your answer would be a five-letter string composed of letters X and N only, e.g. NNXNN.
( ) It is more suitable for complexes larger than about 100 kD.
( ) It requires samples to crystallize.
( ) It can better reveal dynamics of molecular motions.
( ) It involves the calculation of electron-density maps.
( ) It is used to determine the solution structure of proteins.
( ) It is more suitable for complexes larger than about 100 kD.
( ) It requires samples to crystallize.
( ) It can better reveal dynamics of molecular motions.
( ) It involves the calculation of electron-density maps.
( ) It is used to determine the solution structure of proteins.

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
32
Each cycle of PCR doubles the amount of DNA synthesized in the previous cycle. A pair of primers is used to direct the specific synthesis of a desired DNA segment. The following diagram shows the products of PCR amplification after three cycles. The DNA strands of the desired segment are indicated in yellow boxes in the diagram. As shown, at the end of the third cycle, 8 out of 16 strands have the desired length and are boxed. After four more cycles (i.e. at the end of the seventh cycle), how many strands of DNA are expected to be synthesized? How many of them have the desired length? Write down the two numbers and separate them with a comma, e.g. 16,8.
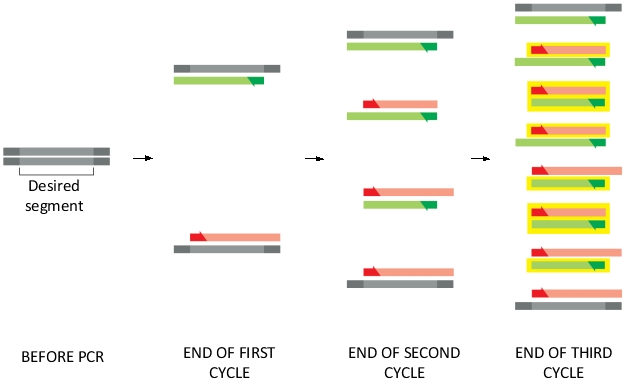


Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
33
In sequence alignments such as those generated by a BLAST search, the significance of the alignment can be presented as an E-value, which specifies how likely it is for a match this good to be found in a database of a given size by chance. You have scanned a large sequence database with a sequence query, hoping to find a significant match. Which one would you look at:
A) a match with an E-value of 1
B) one with an E-value of 0.001
A) a match with an E-value of 1
B) one with an E-value of 0.001

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
34
Indicate whether each of the following descriptions better applies to cDNA libraries (C) or genomic DNA libraries (G). Your answer would be a five-letter string composed of letters C and G only, e.g. CGCGC.
( ) Their production requires use of reverse transcriptase.
( ) They are enriched in protein-coding genes.
( ) Different sequences are represented in them based on their transcription level.
( ) They contain intronic sequences.
( ) They are cell-type specific.
( ) Their production requires use of reverse transcriptase.
( ) They are enriched in protein-coding genes.
( ) Different sequences are represented in them based on their transcription level.
( ) They contain intronic sequences.
( ) They are cell-type specific.

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
35
The EcoRI restriction enzyme recognizes and cuts at the sequence GAATTC. Such an enzyme is expected to cut a typical double-stranded DNA at 1 in every … sites.
A) about 400
B) about 4000
C) about 40,000
D) about 400,000
E) about 4 million
A) about 400
B) about 4000
C) about 40,000
D) about 400,000
E) about 4 million

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
36
Bacterial artificial chromosomes (BACs) …
A) are derived from the E. coli chromosome.
B) are present in high copy numbers per cell.
C) can maintain DNA sequences of hundreds of thousands of nucleotide pairs.
D) are generally not stable.
E) All of the above.
A) are derived from the E. coli chromosome.
B) are present in high copy numbers per cell.
C) can maintain DNA sequences of hundreds of thousands of nucleotide pairs.
D) are generally not stable.
E) All of the above.

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
37
The AgeI restriction enzyme recognizes the sequence ACCGGT and cuts each strand between the first (A) and the second (C) nucleotide. This can be represented as A|CCGGT, with the vertical bar indicating the cleavage position. If a DNA fragment is cut with AgeI at both ends, cutting a plasmid with which of the following enzymes would then allow insertion of the fragment into the plasmid by simple ligation of sticky ends?
A) FseI, with the recognition sequence GGCCGG|CC
B) AatI, with the recognition sequence AGG|CCT
C) BstZI, with the recognition sequence C|GGCCG
D) MroI, with the recognition sequence T|CCGGA
E) None of the above
A) FseI, with the recognition sequence GGCCGG|CC
B) AatI, with the recognition sequence AGG|CCT
C) BstZI, with the recognition sequence C|GGCCG
D) MroI, with the recognition sequence T|CCGGA
E) None of the above

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
38
A researcher has labeled the 5? end of a DNA primer with ³²P radioisotopes. The primer can hybridize to a complementary sequence near the 3? end of a long, spliced mRNA molecule composed of three exons. The mRNA molecule can interact with two RNA-binding proteins: one of them (protein 1) binds to the first exon and the other (protein 2) binds to the second exon. The researcher uses reverse transcriptase to extend the primer in the presence of the mRNA and either of these proteins, then separates the cDNA products by gel electrophoresis, and finally visualizes the labeled DNA bands by autoradiography. The results are presented schematically in the following drawing. Assuming that the reverse transcriptase falls off the template when confronted by bound proteins, which lane (A or B) in the gel corresponds to the reaction in the presence of protein 1? Write down A or B as your answer. 


Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
39
You performed a co-immunoprecipitation experiment to identify cellular proteins associated with protein X. For the experiment, you used antibodies against protein X that were immobilized on beads. You incubated these beads with cell lysates from either wild-type cells or cells lacking the gene encoding protein Y. After washing and eluting the bound protein off the beads, you separated each bound and unbound fraction by SDS-PAGE. Finally, you performed immunoblotting using antibodies against either protein Y or protein Z. The results of the Western blots are presented in the following schematic drawings. According to these results, do you think protein Y enhances (E) or inhibits (I) binding of protein X to protein Z? Write down E or I as your answer. 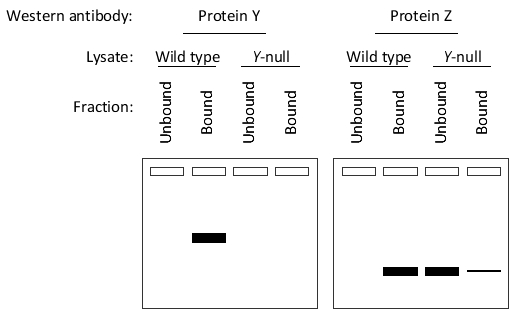


Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
40
Indicate whether ordinary agarose-gel electrophoresis (A), polyacrylamide-gel electrophoresis (P), or pulsed-field gel electrophoresis (F) is used to better separate each of the following DNA molecules. Your answer would be a three-letter string composed of letters A, P, and F only, e.g. APF.
( ) Small DNA oligonucleotides such as hybridization probes and PCR primers
( ) Plasmid DNA
( ) Bacterial chromosomes
( ) Small DNA oligonucleotides such as hybridization probes and PCR primers
( ) Plasmid DNA
( ) Bacterial chromosomes

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
41
You have studied mutations in a transcription regulatory protein from Bacillus subtilis by an in vivo reporter gene assay carried out in Escherichia coli. The cis-regulatory sequence that the protein normally recognizes was inserted upstream of a gene encoding an enzyme that can produce a visible blue product.
A) Their colonies turn blue at permissive (low) temperatures.
B) Their colonies turn blue at permissive (high) temperatures.
C) Their colonies turn blue at nonpermissive (low) temperatures.
D) Their colonies turn blue at nonpermissive (high) temperatures.
E) coli cells were then transformed with the reporter construct along with the gene encoding one of the various mutant forms of the transcription regulatory protein. Since the transcription regulatory protein is an inhibitor, the colonies turn blue only when the mutant protein is nonfunctional and unable to inhibit gene expression. Interestingly, you have identified a number of "cold-sensitive" loss-of-function mutations that show the mutant phenotype at 22°C but not at 37°C. Which of the following correctly describes the phenotype of these cold-sensitive mutants?
A) Their colonies turn blue at permissive (low) temperatures.
B) Their colonies turn blue at permissive (high) temperatures.
C) Their colonies turn blue at nonpermissive (low) temperatures.
D) Their colonies turn blue at nonpermissive (high) temperatures.
E) coli cells were then transformed with the reporter construct along with the gene encoding one of the various mutant forms of the transcription regulatory protein. Since the transcription regulatory protein is an inhibitor, the colonies turn blue only when the mutant protein is nonfunctional and unable to inhibit gene expression. Interestingly, you have identified a number of "cold-sensitive" loss-of-function mutations that show the mutant phenotype at 22°C but not at 37°C. Which of the following correctly describes the phenotype of these cold-sensitive mutants?

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
42
Consider a biochemical pathway in petal cells of a plant, in which a white precursor is turned into a final red pigment through a yellow and an orange intermediate by the activity of three enzymes encoded by genes A, B, and C. The petal color in plants harboring functional (capital letter) or nonfunctional (lowercase letter) alleles of these genes is presented in the following table. What do you think is the order in which the enzymes act in the pathway? Your answer would be a three-letter string composed of letters A to C, e.g. BCA.



Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
43
Indicate whether each of the following descriptions better applies to forward (F) or reverse (R) genetics. Your answer would be a four-letter string composed of letters F and R only, e.g. RFFF.
( ) It involves new gene discovery by genetic screens.
( ) It may also be called genome editing.
( ) In this approach, mutations are introduced deterministically in a known gene.
( ) It is involved in creating knockout mice lacking a particular gene.
( ) It involves new gene discovery by genetic screens.
( ) It may also be called genome editing.
( ) In this approach, mutations are introduced deterministically in a known gene.
( ) It is involved in creating knockout mice lacking a particular gene.

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
44
Two loci on a human chromosome are about 1,500,000 nucleotide pairs apart and are found to recombine in about 2% of gametes during gametogenesis. Consistently, each genetic map unit (which corresponds to 1% recombination) in humans is equivalent to a physical distance of about … nucleotide pairs.
A) 30 million
B) 7.5 million
C) 3 million
D) 0.75 million
E) 0.3
A) 30 million
B) 7.5 million
C) 3 million
D) 0.75 million
E) 0.3

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
45
A piece of DNA has been sequenced by automated dideoxy sequencing, in which each dideoxyribonucleoside triphosphate was labeled with a fluorescent tag of a different color. The data corresponding to a small segment with the sequence 5′-TGCCACA-3′ is shown in the following diagram. Knowing that ddGTP has been labeled with green fluorescence, what color do you think was the fluorescent tag used to label ddTTP in the sequencing experiment? 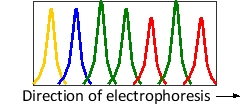
A) Blue
B) Green
C) Red
D) Yellow

A) Blue
B) Green
C) Red
D) Yellow

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
46
Susceptibility to drug and alcohol addiction is partially genetic. For example, the human dopamine receptor DRD2 is associated with cocaine and alcohol abuse; individuals carrying an A nucleotide, instead of the more common C nucleotide, at a certain position within the sixth intron of the DRD2 gene are at a significantly increased risk of abuse. This variation in sequence is an example of …
A) a SNP.
B) a CNV.
C) an indel.
D) epistasis.
E) None of the above.
A) a SNP.
B) a CNV.
C) an indel.
D) epistasis.
E) None of the above.

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
47
If two genomic polymorphic sites were randomly and independently associated with each other, one would expect the frequency of observing a combination of their allelic forms in an individual to be equal to the multiplication product of the probability of the alleles occurring separately. In many cases, however, this is not true. The degree of deviation from random association can be quantified and summarized in diagrams such as the one below. In this diagram, eight SNPs in a genomic region are shown and their pairwise deviation from random association is color-coded by different shades of red, such that the darkest red shade indicates the furthest deviation from random association. In contrast, those pairs that associate randomly are colored white. For example, SNPs A and F seem to associate randomly as indicated by the corresponding box marked with black borders. From this diagram, two haplotype blocks can be readily detected. What subset of the shown SNPs is not part of any of the two blocks? Write down the letter(s) as your answer, in alphabetical order, e.g. CEH. 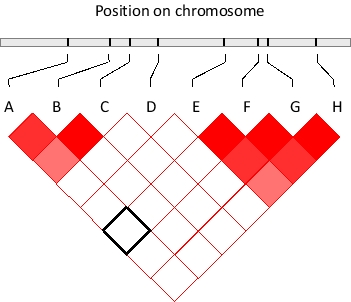


Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
48
A piece of DNA has been sequenced by Ion Torrent™ sequencing. The result corresponding to a six-nucleotide segment of this DNA is presented in the graph below, in which pH change is recorded after the addition of each of the four nucleotide triphosphates in cycles. What is the sequence of this segment? All sequences are written from 5′ to 3′. 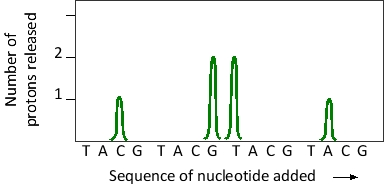
A) TACGTA
B) CGTACG
C) CCGTAA
D) CGGTTA
E) ATTGGC

A) TACGTA
B) CGTACG
C) CCGTAA
D) CGGTTA
E) ATTGGC

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
49
In a bacterium that is normally cylindrical in shape, loss of either of two genes A and B results in round cells. Additionally, gain-of-function mutations in gene A can rescue the phenotype of gene B loss-of-function mutants. What phenomenon best describes this observation? The product of which gene is more likely to function upstream of the other one in determining cell shape in this bacterium?
A) Suppression; gene A
B) Suppression; gene B
C) Complementation; gene A
D) Complementation; gene B
A) Suppression; gene A
B) Suppression; gene B
C) Complementation; gene A
D) Complementation; gene B

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
50
A portion of RNA-seq data obtained from two tissue samples is plotted in the following schematic diagram. Which region (A to E) corresponds to an alternative exon? 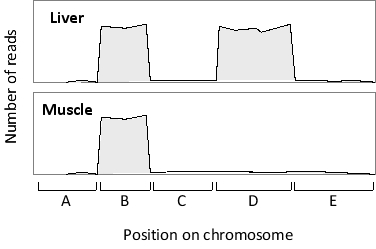


Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
51
For a complementation test to work, the mutations under study must be…
A) recessive.
B) dominant.
C) gain-of-function.
D) null.
E) conditional.
A) recessive.
B) dominant.
C) gain-of-function.
D) null.
E) conditional.

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
52
In Sanger sequencing, does each labeled DNA molecule have a fluorescently labeled dideoxyribonucleotide at its 5′ end or its 3′ end? Do the labeled nucleotides lack the 2′ or 3′ hydroxyl group?
A) 3′ end; 2′ hydroxyl group only
B) 3′ end; 3′ hydroxyl group only
C) 5′ end; 2′ hydroxyl group only
D) 5′ end; 3′ hydroxyl group only
E) 3′ end; both hydroxyl groups
A) 3′ end; 2′ hydroxyl group only
B) 3′ end; 3′ hydroxyl group only
C) 5′ end; 2′ hydroxyl group only
D) 5′ end; 3′ hydroxyl group only
E) 3′ end; both hydroxyl groups

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
53
You would like to clone a gene by PCR. The sequence of the two ends of the coding strand for the gene is provided below. Which following pair of primers would direct the amplification of this gene? All sequences are written from 5? to 3?.
ATGAAATCTACGTTTCAC……CCCCCAGTACCCCCCTTA
A) ATGAAATCTACGTTTCAC ; CCCCCAGTACCCCCCTTA
B) ATGAAATCTACGTTTCAC ; TAAGGGGGGTACTGGGGG
C) TACTTTAGATGCAAAGTG; ATTCCCCCCATGACCCCC
D) TACTTTAGATGCAAAGTG ; CCCCCAGTACCCCCCTTA
E) GTGAAACGTAGATTTCAT; ATTCCCCCCATGACCCCC
ATGAAATCTACGTTTCAC……CCCCCAGTACCCCCCTTA
A) ATGAAATCTACGTTTCAC ; CCCCCAGTACCCCCCTTA
B) ATGAAATCTACGTTTCAC ; TAAGGGGGGTACTGGGGG
C) TACTTTAGATGCAAAGTG; ATTCCCCCCATGACCCCC
D) TACTTTAGATGCAAAGTG ; CCCCCAGTACCCCCCTTA
E) GTGAAACGTAGATTTCAT; ATTCCCCCCATGACCCCC

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
54
Loss-of-function mutations in either imp or yfgL genes in Escherichia coli result in an increased permeability of the outer membrane to small molecules and therefore confer antibiotic sensitivity, as the antibiotics can enter the cell more readily. Interestingly, double mutants, in which both genes are defective, are resistant to antibiotics. Similar to the wild-type strain, but unlike the single mutants, they can grow on media containing antibiotics. This is an example of …
A) synthetic lethality.
B) complementation.
C) synergy.
D) a recessive mutation.
E) None of the above.
A) synthetic lethality.
B) complementation.
C) synergy.
D) a recessive mutation.
E) None of the above.

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
55
A protein made from an expression vector can be so plentiful as to account for about 10% of total cell protein. You would like to produce a protein of interest that can be expressed at such a level in Escherichia coli. You plan to induce the expression and collect the cells when cell density in the culture medium reaches about 10? cells/mL. Proteins make up about 15% of total cell weight, and each E. coli cell typically has a mass of about 1 picogram (i.e. 10-¹² g). Assuming that almost 50% of the protein is lost during purification, how many liters of culture should you prepare in order to obtain 15 milligrams of pure protein? Write down your answer in liters, with two decimal places and without units, e.g. 15.00.

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
56
You have isolated five mutations (1 to 5) in the yeast Saccharomyces cerevisiae that make the haploid cells unable to grow in the absence of histidine. Each haploid mutant can be mated with any of the other ones, forming diploid cells that either can (+) or cannot (-) grow in the absence of histidine, as indicated in the following complementation table. How many complementation groups do these mutations represent? Each complementation group typically corresponds to a separate gene. 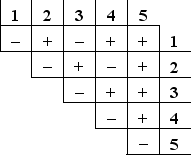
A) 1
B) 2
C) 3
D) 4
E) 5

A) 1
B) 2
C) 3
D) 4
E) 5

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
57
The wild-type nematode Caenorhabditis elegans has a bent body with the shape of a wave. In the presence of increasing concentrations of ethanol, however, the body bend is progressively diminished. Loss-of-function mutations in SLO-1, a gene that codes for an ion channel, confer ethanol resistance and allow the worm to maintain its body bend up to higher ethanol concentrations. The channel function is defined based on the ability of the channel to conduct ions across the membrane. According to these observations, would you expect the channel to open or close in the presence of ethanol in wild-type worms? Would a gain-of-function mutation in SLO-1 tend to increase or decrease body bend?
A) Open; increase
B) Open; decrease
C) Close; increase
D) Close; decrease
A) Open; increase
B) Open; decrease
C) Close; increase
D) Close; decrease

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
58
Indicate whether each of the following descriptions better applies to Illumina sequencing (I), Ion Torrent™ sequencing (T), or both sequencing technologies (B). Your answer would be a four-letter string composed of letters B, I, and T only, e.g. IBBB.
( ) It uses fluorescently labeled nucleotides.
( ) It uses PCR-generated copies of DNA.
( ) It is a second-generation sequencing technology and employs a cyclic wash-and-measure paradigm.
( ) It relies on the fidelity of DNA polymerase for its accuracy.
( ) It uses fluorescently labeled nucleotides.
( ) It uses PCR-generated copies of DNA.
( ) It is a second-generation sequencing technology and employs a cyclic wash-and-measure paradigm.
( ) It relies on the fidelity of DNA polymerase for its accuracy.

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
59
There are six possible reading frames for any double-stranded DNA. In the following schematic drawing of a genomic region in Escherichia coli, stop codons in each of these frames are indicated by black bars. In which reading frame do you think the open reading frame (ORF) is translated in this region? Write down the frame number as your answer, e.g. 2.
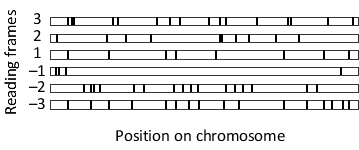


Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
60
The p53 gene is an important tumor suppressor gene in our genome. It codes for a transcription regulatory protein that can assemble into tetramers. A number of mutations in p53 render the protein inactive. These mutant protein molecules form mixed tetramers with wild-type p53 proteins and, in this case, the tetramer is inactive. The mutations can therefore be categorized as …
A) null.
B) gain-of-function.
C) recessive lethal.
D) dominant-negative.
E) suppressor.
A) null.
B) gain-of-function.
C) recessive lethal.
D) dominant-negative.
E) suppressor.

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
61
You have created transgenic mice that carry the gene encoding green fluorescent protein (GFP) under the control of a ubiquitous promoter. The GFP-encoding gene, however, is inactive unless recombination by Cre removes a blocking sequence between the promoter and the gene. These mice have been mated with those harboring the Cre recombinase under the control of the insulin promoter. The Cre recombinase is fused to the estrogen receptor, which keeps the fusion in the cytosol unless it binds to tamoxifen, in which case it allows nuclear entry. After mating, you select adult mice harboring both of these transgenes and inject them with either tamoxifen or water. Which of the following would you expect to observe later as a result when you examine brain and pancreas tissue samples under a fluorescence microscope?
A) GFP signal is observed in the brain in the presence of tamoxifen only.
B) GFP signal is observed in the brain in the absence of tamoxifen only.
C) GFP signal is observed in the pancreas in the presence of tamoxifen only.
D) GFP signal is observed in the pancreas in the absence of tamoxifen only.
E) GFP signal is observed in the brain in either the presence or the absence of tamoxifen.
A) GFP signal is observed in the brain in the presence of tamoxifen only.
B) GFP signal is observed in the brain in the absence of tamoxifen only.
C) GFP signal is observed in the pancreas in the presence of tamoxifen only.
D) GFP signal is observed in the pancreas in the absence of tamoxifen only.
E) GFP signal is observed in the brain in either the presence or the absence of tamoxifen.

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
62
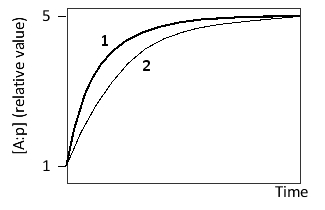
Consider a transcription regulatory protein (A) that can bind to a promoter (p). After reaching an initial steady state, the concentration of A is suddenly increased tenfold. The following graph shows the formation of A:p over time after this change for two systems, one of which is described by higher k?ff and k?n values compared to the other. Which curve (1 or 2) corresponds to this system? Write down 1 or 2 as your answer.


Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
63
Consider the binding of a transcription regulatory protein (A) to a promoter DNA (p) according to the following scheme.
A + p?A:p
At steady state, which of the following formulae correctly yields the bound promoter fraction, i.e. [A:p]/([A:p] + [p])? The association constant is designated as K.
A) [A]/(K + [A])
B) K[A]/(1 + K[A])
C) K[A]/(K + [A])
D) [A]/(1 + K[A])
A + p?A:p
At steady state, which of the following formulae correctly yields the bound promoter fraction, i.e. [A:p]/([A:p] + [p])? The association constant is designated as K.
A) [A]/(K + [A])
B) K[A]/(1 + K[A])
C) K[A]/(K + [A])
D) [A]/(1 + K[A])

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
64
Consider a protein composed of only 21 amino acids encoded by a short ORF that is located between two known genes in a mammalian genome. Due to its length, the ORF had not been annotated as a gene. Which of the following methods can demonstrate that it is indeed a gene that is actively translated to protein?
A) RNA-seq
B) RT-PCR
C) DNA microarray analysis
D) ChIP-seq
E) Ribosome profiling
A) RNA-seq
B) RT-PCR
C) DNA microarray analysis
D) ChIP-seq
E) Ribosome profiling

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
65
RNA-seq and ribosome profiling experiments have been carried out on the same cells. The following simplified graphs show the results for the same genomic region containing a gene with three exons. Which graph (1 or 2) do you think corresponds to the RNA-seq results? What feature is represented by the arrow? 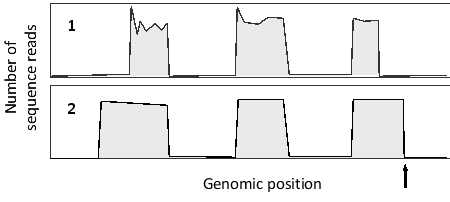
A) Graph 1; stop codon
B) Graph 1; polyadenylation site
C) Graph 2; stop codon
D) Graph 2; polyadenylation site

A) Graph 1; stop codon
B) Graph 1; polyadenylation site
C) Graph 2; stop codon
D) Graph 2; polyadenylation site

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
66
When a promoter p is not saturated with bound regulatory protein A, doubling the total concentration of A would …
A) increase steady-state [A:p] exactly twofold.
B) increase steady-state [A:p] over twofold.
C) increase steady-state [A:p] less than twofold.
D) decrease steady-state [A:p] approximately twofold.
E) decrease steady-state [A:p] over twofold.
A) increase steady-state [A:p] exactly twofold.
B) increase steady-state [A:p] over twofold.
C) increase steady-state [A:p] less than twofold.
D) decrease steady-state [A:p] approximately twofold.
E) decrease steady-state [A:p] over twofold.

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
67
You have used a reporter gene system to investigate the contribution to gene expression of three cis-regulatory DNA sequences (A to C) from a key developmental gene. Reporter gene expression from constructs containing all three or only a subset of the regulatory sequences is summarized in the diagram below. According to these findings, indicate whether each of the following conclusions is (Y) or is not (N) acceptable. Your answer would be a four-letter string composed of letters Y and N only, e.g. NNNN.
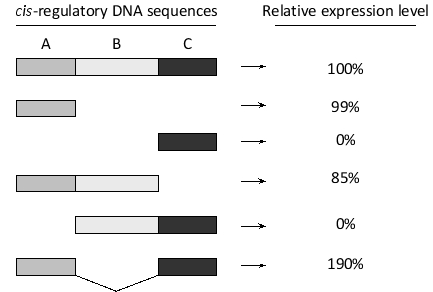
( ) Region B is likely to normally bind an activator and turn the gene on.
( ) Region C is sufficient for activation of gene expression.
( ) Region A is necessary for activation of gene expression.
( ) Region C is necessary for activation of gene expression.

( ) Region B is likely to normally bind an activator and turn the gene on.
( ) Region C is sufficient for activation of gene expression.
( ) Region A is necessary for activation of gene expression.
( ) Region C is necessary for activation of gene expression.

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
68
Consider a transcription regulatory protein (A) that can bind to the promoter (p) of a target gene. After reaching an initial steady state, the concentration of A is suddenly increased tenfold. The following graph shows the formation of A:p over time after this change. What is the slope of the curve at time zero? 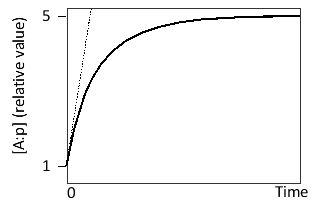
A) 2 kₒff
B) 2 kₒn
C) 5 kₒff
D) 5 kₒn
E) 9 kₒff

A) 2 kₒff
B) 2 kₒn
C) 5 kₒff
D) 5 kₒn
E) 9 kₒff

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
69
Everything else being equal, if the mean lifetime of a protein is doubled, its concentration at steady state would …
A) increase approximately twofold.
B) increase less than twofold.
C) increase over twofold.
D) decrease approximately twofold.
E) decrease less than twofold.
A) increase approximately twofold.
B) increase less than twofold.
C) increase over twofold.
D) decrease approximately twofold.
E) decrease less than twofold.

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
70
Sort the following steps in the common procedure to create transgenic plants. Your answer would be a four-letter string composed of letters A to D, e.g. DABC.
(A) Callus growth
(B) Agrobacterium infection
(C) Shoot/root induction
(D) Removal of leaf tissue from a plant
(A) Callus growth
(B) Agrobacterium infection
(C) Shoot/root induction
(D) Removal of leaf tissue from a plant

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
71
You mated male and female heterozygous Oct4 knockout mice to study the function of this important gene. Among the progeny, one-third were homozygous wild type and the rest were heterozygous. You then repeated the cross, but this time you genotyped several embryos at different stages of early development and grouped them as either homozygous wild type (+/+), heterozygous (+/-), or homozygous null (-/-). You have summarized your results in the following table, in which the number of embryos in different groups and at different developmental stages are presented. According to these results, complete loss of Oct4 is … 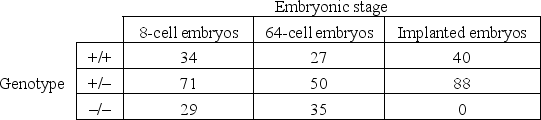
A) NOT lethal.
B) synthetic lethal.
C) zygotic lethal.
D) embryonic lethal.

A) NOT lethal.
B) synthetic lethal.
C) zygotic lethal.
D) embryonic lethal.

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
72
Which of the following can limit the use of RNA interference in studying gene expression and function?
A) RNAi does not efficiently inactivate all genes.
B) Certain tissues are resistant to the action of RNAi.
C) Off-target effects are common in RNAi treatments.
D) All of the above.
A) RNAi does not efficiently inactivate all genes.
B) Certain tissues are resistant to the action of RNAi.
C) Off-target effects are common in RNAi treatments.
D) All of the above.

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
73
Which of the following methods provides the most sensitive and accurate way of measuring and comparing mRNA levels of a set of genes in two different tissues. The genes are expressed at relatively low levels in these tissues, and do not necessarily code for protein.
A) RNA-seq
B) RT-PCR
C) DNA microarray analysis
D) ChIP-seq
E) Ribosome profiling
A) RNA-seq
B) RT-PCR
C) DNA microarray analysis
D) ChIP-seq
E) Ribosome profiling

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
74
The results of two sets of chromatin immunoprecipitation experiments on two cell types (1 and 2) are shown in the following simplified graphs for the same genomic region containing the first two exons of a gene. Antibodies against a transcription regulatory protein (X) or against a subunit of RNA polymerase II were used in the ChIP-seq experiments, as indicated. Do you think the protein is a transcriptional activator or repressor? The results from a parallel RNA-seq experiment are provided in the third graph. Which profile (a or b) in the graph would you expect to correspond to cell type 1? 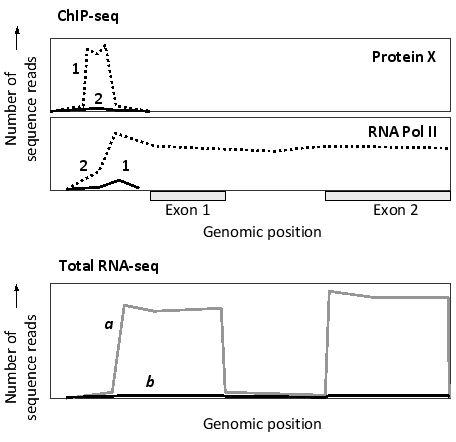
A) Activator; profile a
B) Repressor; profile a
C) Activator; profile b
D) Repressor; profile b

A) Activator; profile a
B) Repressor; profile a
C) Activator; profile b
D) Repressor; profile b

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
75
In the following schematic graph, the fraction of promoters (px) bound to the regulatory protein A is plotted at various concentrations of A. Which of the following quantities represents X in the graph? ![<strong>In the following schematic graph, the fraction of promoters (px) bound to the regulatory protein A is plotted at various concentrations of A. Which of the following quantities represents X in the graph? </strong> A) [Total pₓ] B) [pₓ] C)kₒn / kₒff D) kₒff / kₒn E)kon[px] / koff](https://storage.examlex.com/TB1030/11ea33de_1b61_023a_bddc_7be2fd36698f_TB1030_00.jpg)
A) [Total pₓ]
B) [pₓ]
C)kₒn / kₒff
D) kₒff / kₒn
E)kon[px] / koff
![<strong>In the following schematic graph, the fraction of promoters (px) bound to the regulatory protein A is plotted at various concentrations of A. Which of the following quantities represents X in the graph? </strong> A) [Total pₓ] B) [pₓ] C)kₒn / kₒff D) kₒff / kₒn E)kon[px] / koff](https://storage.examlex.com/TB1030/11ea33de_1b61_023a_bddc_7be2fd36698f_TB1030_00.jpg)
A) [Total pₓ]
B) [pₓ]
C)kₒn / kₒff
D) kₒff / kₒn
E)kon[px] / koff

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
76
A pool of five barcoded yeast mutants, each deleted for a different gene (genes A to E), has been grown under three different conditions (1 to 3). After the growth phase was completed, genomic DNA was isolated from the entire pool and the relative amount of each mutant was determined by quantifying the amount of DNA containing each barcode. The results are presented in the following graph. Which gene (A to E) is essential under condition 1 but not under conditions 2 or 3? 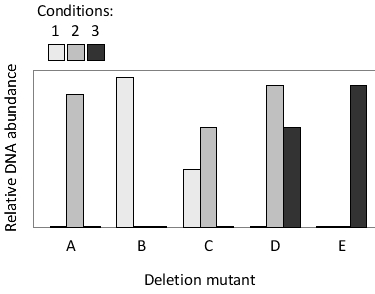


Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
77
The following schematic graph shows the result of two quantitative RT-PCR experiments. Total mRNA from two tissue samples (1 and 2) was isolated and subjected to RT-PCR using primers designed to amplify a tissue-specific gene. Added to the reaction was a fluorescent dye that fluoresces only when bound to double-stranded DNA. According to the graph, which tissue has a higher level of this mRNA? By how much? 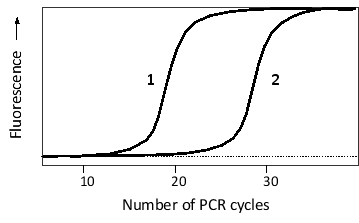
A) About 2-fold higher in tissue 1
B) About 1000-fold higher in tissue 1
C) About 2-fold higher in tissue 2
D) About 1000-fold higher in tissue 2

A) About 2-fold higher in tissue 1
B) About 1000-fold higher in tissue 1
C) About 2-fold higher in tissue 2
D) About 1000-fold higher in tissue 2

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
78
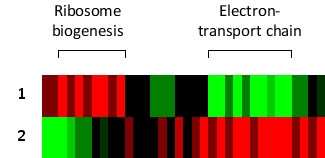
You have grown cultures of the yeast Saccharomyces cerevisiae in a complex medium alone or supplemented with either glucose or ethanol. In the presence of glucose, available energy is abundant and the cells can therefore grow by fermenting glucose. In the presence of ethanol, however, the cells switch to a mostly aerobic respiration to meet their energy demands. They also shut down many of their anabolic pathways and activate catabolic pathways to produce more energy to better survive under these conditions. After the growth phase, you extract total mRNA from the cells, convert them to cDNA, and label them with a fluorescent molecule. You then hybridize the cDNAs to a yeast DNA microarray. Part of the results obtained by cluster analysis after fluorescent measurements is summarized in the following simplified diagram. Here, red and green indicate increase and decrease, respectively, in expression of a gene compared to that in the nonsupplemented medium. Which row (1 or 2) do you think corresponds to the glucose-supplemented medium?


Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
79
In which of the following organisms is systemic RNAi possible through feeding the animal with the RNA?
A) Caenorhabditis elegans
B) Drosophila melanogaster
C) Mus musculus
D) Homo sapiens
E) All of the above
A) Caenorhabditis elegans
B) Drosophila melanogaster
C) Mus musculus
D) Homo sapiens
E) All of the above

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck
80
Which of the following is NOT correct regarding both Cas9 and EcoRI?
A) They are both endonucleases.
B) They both create double-strand breaks in DNA.
C) They both recognize their target sequences with the help of guide RNAs.
D) They are both part of bacterial defense mechanisms against foreign DNA.
E) They are both greatly useful in manipulating DNA and studying gene expression and function.
A) They are both endonucleases.
B) They both create double-strand breaks in DNA.
C) They both recognize their target sequences with the help of guide RNAs.
D) They are both part of bacterial defense mechanisms against foreign DNA.
E) They are both greatly useful in manipulating DNA and studying gene expression and function.

Unlock Deck
Unlock for access to all 95 flashcards in this deck.
Unlock Deck
k this deck


