Deck 8: Recombinant Dna Technology and Molecular Cloning
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/85
Play
Full screen (f)
Deck 8: Recombinant Dna Technology and Molecular Cloning
1
Following the injection of a linear phage DNA into E. coli host cells, the phage genome circularizes by
A) recombination at attachment (Att) sites.
B) joining of the attachment (Att) sites.
C) recombination at cohesive (cos) sites.
D) joining of the cohesive (cos) sites.
A) recombination at attachment (Att) sites.
B) joining of the attachment (Att) sites.
C) recombination at cohesive (cos) sites.
D) joining of the cohesive (cos) sites.
joining of the cohesive (cos) sites.
2
When E. coli host strain K-12 is infected by phage lambda K, the phage DNA is
A) recognized as foreign and cleaved by a specific restriction endonuclease.
B) recognized as foreign and marked with a methylation pattern.
C) not recognized as foreign because it has the same methylation pattern as the E. coli genome
D) not recognized as foreign because of the phage restriction and modification system.
A) recognized as foreign and cleaved by a specific restriction endonuclease.
B) recognized as foreign and marked with a methylation pattern.
C) not recognized as foreign because it has the same methylation pattern as the E. coli genome
D) not recognized as foreign because of the phage restriction and modification system.
not recognized as foreign because it has the same methylation pattern as the E. coli genome
3
Recombinant DNA technology became possible with the discovery of which of the following enzymes?
A) terminal deoxynucleotidyl transferase
B) restriction endonucleases and DNA ligase
C) DNA polymerase and DNA ligase
D) reverse transcriptase
A) terminal deoxynucleotidyl transferase
B) restriction endonucleases and DNA ligase
C) DNA polymerase and DNA ligase
D) reverse transcriptase
restriction endonucleases and DNA ligase
4
Type II restriction endonucleases are useful in recombinant DNA research primarily because
A) they cut both strands of DNA at a specific recognition site.
B) they can distinguish between genomic and cDNA.
C) they can join two pieces of DNA by forming phosphodiester bonds.
D) they cleave DNA at a distance from the recognition site.
A) they cut both strands of DNA at a specific recognition site.
B) they can distinguish between genomic and cDNA.
C) they can join two pieces of DNA by forming phosphodiester bonds.
D) they cleave DNA at a distance from the recognition site.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
5
Which statement is not true about DNA binding and cleavage by type II restriction endonucleases?
A) the recognition process triggers a conformational change in both the enzyme and the DNA.
B) the first contact with DNA is specific.
C) the target site is located by sliding, hopping, and jumping.
D) the enzyme interacts directly with the nitrogenous bases.
A) the recognition process triggers a conformational change in both the enzyme and the DNA.
B) the first contact with DNA is specific.
C) the target site is located by sliding, hopping, and jumping.
D) the enzyme interacts directly with the nitrogenous bases.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
6
What is suggested by the fact that type II restriction endonuclease recognition sequences are usually palindromic?
A) A particular restriction endonuclease can cleave DNA at several different sites.
B) Restriction endonucleases must cleave DNA in a multistep process.
C) Restriction endonucleases function as dimers.
D) Restriction endonucleases recognize the sugar-phosphate backbone of DNA.
A) A particular restriction endonuclease can cleave DNA at several different sites.
B) Restriction endonucleases must cleave DNA in a multistep process.
C) Restriction endonucleases function as dimers.
D) Restriction endonucleases recognize the sugar-phosphate backbone of DNA.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
7
Which statement is not true about DNA ligase?
A) It requires ATP as a cofactor.
B) It joins two pieces of DNA by forming phosphodiester bonds.
C) It can circularize a linear DNA with blunt ends.
D) It can only ligate fragments of DNA with sticky (complementary) ends.
A) It requires ATP as a cofactor.
B) It joins two pieces of DNA by forming phosphodiester bonds.
C) It can circularize a linear DNA with blunt ends.
D) It can only ligate fragments of DNA with sticky (complementary) ends.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
8
In the context of recombinant DNA technology, plasmids are used as vectors. This means
That plasmids
A) allow for transfer of foreign DNA molecules into a host cell.
B) allow for infection and lysis of host cells.
C) serve as host cells for amplification of foreign DNA molecules.
D) function as artificial chromosomes in host cells.
That plasmids
A) allow for transfer of foreign DNA molecules into a host cell.
B) allow for infection and lysis of host cells.
C) serve as host cells for amplification of foreign DNA molecules.
D) function as artificial chromosomes in host cells.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
9
Why is E. coli so widely used as a host organism in which to clone genes?
A) E. coli has a rapid growth rate.
B) Growing cultures of E. coli cells is relatively easy.
C) Many different E. coli strains with distinct and useful properties are available.
D) All of the above
A) E. coli has a rapid growth rate.
B) Growing cultures of E. coli cells is relatively easy.
C) Many different E. coli strains with distinct and useful properties are available.
D) All of the above

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
10
In recombinant DNA technology, the term competency refers to
A) the ability of DNA ends to be ligated
B) the ability of an enzyme to be activated by ATP
C) the ability of cells to take up foreign DNA
D) the ability of a medium to select for transformed cells
A) the ability of DNA ends to be ligated
B) the ability of an enzyme to be activated by ATP
C) the ability of cells to take up foreign DNA
D) the ability of a medium to select for transformed cells

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
11
Which of the following vectors can accommodate the largest DNA insert?
A) plasmid
B) cosmid
C) phage
D) pUC18
A) plasmid
B) cosmid
C) phage
D) pUC18

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
12
You have ligated a foreign DNA fragment into the EcoRI site in the multiple cloning site of pUC18. You then transform host E. coli with the ligation mixture. Some of the bacterial colonies growing on the nutrient agar plate that contains ampicillin and X-gal are white and some are blue. Bacterial colonies that are blue
A) contain recombinant plasmid DNA
B) contain nonrecombinant plasmid DNA
C) are untransformed bacteria
D) have hydrolyzed ampicillin to form a colored compound
A) contain recombinant plasmid DNA
B) contain nonrecombinant plasmid DNA
C) are untransformed bacteria
D) have hydrolyzed ampicillin to form a colored compound

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
13
To separate acidic proteins from basic proteins in a crude cellular lysate you could use
A) gel filtration chromatography
B) antibody-affinity chromatography
C) ion-exchange chromatography
D) A and C
A) gel filtration chromatography
B) antibody-affinity chromatography
C) ion-exchange chromatography
D) A and C

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
14
In order to function, yeast artificial chromosome (YAC) vectors must include the following:
A) an origin of replication
B) a centromere and telomeres
C) growth selectable markers
D) all of the above
A) an origin of replication
B) a centromere and telomeres
C) growth selectable markers
D) all of the above

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
15
In red-white selection for recombinant yeast artificial chromosome (YAC) vectors in yeast cells, a red colony contains
A) recombinant YAC vector DNA.
B) nonrecombinant YAC vector DNA.
C) no YAC vector DNA.
D) circular YAC vector DNA
A) recombinant YAC vector DNA.
B) nonrecombinant YAC vector DNA.
C) no YAC vector DNA.
D) circular YAC vector DNA

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
16
How does a genomic library differ from a gene clone?
A) A genomic library contains many different sequences that represent the entire genome of an organism; a gene clone contains one type of sequence.
B) A genomic library contains one type of sequence; a gene clone contains many different sequences that represent the entire genome of an organism.
C) A genomic library is much smaller than a gene clone.
D) A genomic library is sequence information stored in silico (i.e., in a computer); a gene clone is an actual sequence of DNA in a plasmid vector.
A) A genomic library contains many different sequences that represent the entire genome of an organism; a gene clone contains one type of sequence.
B) A genomic library contains one type of sequence; a gene clone contains many different sequences that represent the entire genome of an organism.
C) A genomic library is much smaller than a gene clone.
D) A genomic library is sequence information stored in silico (i.e., in a computer); a gene clone is an actual sequence of DNA in a plasmid vector.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
17
What aspect of a common recombinant DNA technology tool makes direct use of knowledge acquired from studies of retrovirus replication?
A) Using restriction endonucleases to cleave DNA
B) Complementary DNA (cDNA) synthesis
C) Introducing DNA into bacterial cells by transformation
D) Using DNA ligase to join two pieces of DNA
A) Using restriction endonucleases to cleave DNA
B) Complementary DNA (cDNA) synthesis
C) Introducing DNA into bacterial cells by transformation
D) Using DNA ligase to join two pieces of DNA

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
18
Reverse transcriptase synthesizes
A) DNA from DNA template
B) DNA from an RNA template
C) RNA from a DNA template
D) RNA from an RNA template
A) DNA from DNA template
B) DNA from an RNA template
C) RNA from a DNA template
D) RNA from an RNA template

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
19
A cDNA library contains
A) DNA fragments that represent the entire set of genes in the genome of an organism.
B) DNA fragments that represent the coding regions of expressed genes from a specific tissue, cell type or developmental stage.
C) DNA fragments that represent the complete sequence of expressed genes from a specific tissue, cell type or developmental stage.
D) RNA fragments that represent the coding regions of expressed genes from a specific tissue, cell type or developmental stage.
A) DNA fragments that represent the entire set of genes in the genome of an organism.
B) DNA fragments that represent the coding regions of expressed genes from a specific tissue, cell type or developmental stage.
C) DNA fragments that represent the complete sequence of expressed genes from a specific tissue, cell type or developmental stage.
D) RNA fragments that represent the coding regions of expressed genes from a specific tissue, cell type or developmental stage.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
20
How can an amino acid sequence be used to design a gene-specific hybridization probe?
A) Synthesize a portion of a polypeptide chain based on the amino acid sequence of the full protein, and use this polypeptide chain as a probe.
B) Purify a protein that contains this amino acid sequence, digest it with proteases that cleave at specific sites, and use one of these peptide fragments as a probe.
C) Synthesize all possible nucleotide sequences long enough to encode the amino acid sequence and thereby find one sequence that will encode the amino acid sequence.
D) Synthesize a set of nucleotide sequences that could encode the amino acid sequence, and this set of oligonucleotides as a probe.
A) Synthesize a portion of a polypeptide chain based on the amino acid sequence of the full protein, and use this polypeptide chain as a probe.
B) Purify a protein that contains this amino acid sequence, digest it with proteases that cleave at specific sites, and use one of these peptide fragments as a probe.
C) Synthesize all possible nucleotide sequences long enough to encode the amino acid sequence and thereby find one sequence that will encode the amino acid sequence.
D) Synthesize a set of nucleotide sequences that could encode the amino acid sequence, and this set of oligonucleotides as a probe.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
21
Which of the following sequences is in the correct order for one cycle of PCR?
A) Denature DNA; primer extension, anneal primers.
B) Primer extension; anneal primers, denature DNA.
C) Anneal primers; denature DNA; primer extension.
D) Denature DNA; anneal primers; primer extension.
A) Denature DNA; primer extension, anneal primers.
B) Primer extension; anneal primers, denature DNA.
C) Anneal primers; denature DNA; primer extension.
D) Denature DNA; anneal primers; primer extension.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
22
In a single PCR cycle consisting of 30 seconds at 95C, 1 min at 55C, and 1 min at 72C, what is happening in the step run at 55C?
A) The DNA to be amplified is being denatured.
B) Primers are being denatured.
C) DNA polymerase is extending new DNA from the primers.
D) Primers are annealing to the DNA to be amplified.
A) The DNA to be amplified is being denatured.
B) Primers are being denatured.
C) DNA polymerase is extending new DNA from the primers.
D) Primers are annealing to the DNA to be amplified.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
23
Which of the following enzymes is involved in the PCR process?
A) DNA ligase
B) reverse transcriptase
C) DNA polymerase
D) DNA primase
A) DNA ligase
B) reverse transcriptase
C) DNA polymerase
D) DNA primase

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
24
Phosphorus-32 (32P) is commonly used for labeling
A) dNTPs and NTPs
B) amino acids
C) nitrogenous bases
D) chloramphenicol
A) dNTPs and NTPs
B) amino acids
C) nitrogenous bases
D) chloramphenicol

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
25
What does it mean when some bands on an autoradiograph are darker than others?
A) The concentration of the labeled macromolecule in that band is higher.
B) The concentration of the labeled macromolecule in that band is lower.
C) The experiment failed because the bands should be of the same intensity throughout the autoradiograph.
D) The darker bands represent larger labeled macromolecules.
A) The concentration of the labeled macromolecule in that band is higher.
B) The concentration of the labeled macromolecule in that band is lower.
C) The experiment failed because the bands should be of the same intensity throughout the autoradiograph.
D) The darker bands represent larger labeled macromolecules.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
26
To prepare a hybridization probe for screening a library, which labeling technique would be most desirable?
A) Klenow fill-in
B) 5 ' end-labeling
C) random primed labeling
D) 3 'end-labeling
A) Klenow fill-in
B) 5 ' end-labeling
C) random primed labeling
D) 3 'end-labeling

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
27
The appropriate hybridization temperature during library screening is calculated according to the G + C content and the percent homology of the probe to target. The more mismatches there are:
A) the higher the melting temperature (Tm) will be.
B) the lower the melting temperature (Tm) will be.
C) the more stable the heteroduplex will be.
D) the more stringent the wash should be.
A) the higher the melting temperature (Tm) will be.
B) the lower the melting temperature (Tm) will be.
C) the more stable the heteroduplex will be.
D) the more stringent the wash should be.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
28
You have screened a library by nucleic acid hybridization with a probe for a particular gene sequence. In the final phase of library screening, you apply an X-ray film to the membrane. The resulting autoradiogram has two black spots corresponding to two separate bacterial colonies. Which of the following explanations is the most likely?
A) The gene is large and fragmented between two different clones.
B) One of the spots must be an artifact because only one bacterial colony should contain the gene of interest.
C) The library screening was a failure because there should be blue and white colonies on the autoradiogram.
D) The temperature at which the high stringency washes was performed was too high.
A) The gene is large and fragmented between two different clones.
B) One of the spots must be an artifact because only one bacterial colony should contain the gene of interest.
C) The library screening was a failure because there should be blue and white colonies on the autoradiogram.
D) The temperature at which the high stringency washes was performed was too high.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
29
What is the difference between a restriction endonuclease recognition site and a RFLP?
A) Restriction endonuclease recognition sites are any sequence of DNA recognized by a restriction endonuclease; RFLPs are any sequence of RNA amplified by reverse transcriptase and PCR.
B) Restriction endonuclease recognition sites produce DNA fragments that differ in size when the source of the DNA is from different chromosomes; RFLPs are any sequence of RNA amplified by reverse transcriptase and PCR.
C) Restriction endonuclease recognition sites produce DNA fragments that differ in size when the source of the DNA is from different chromosomes; RFLPs are any sequence of DNA recognized by a restriction endonuclease.
D) Restriction endonuclease recognition sites are any sequence of DNA recognized by a restriction endonuclease; RFLPs are DNA fragments produced by restriction endonuclease digestion that differ in size when DNA sequences differ between chromosomes or individuals.
A) Restriction endonuclease recognition sites are any sequence of DNA recognized by a restriction endonuclease; RFLPs are any sequence of RNA amplified by reverse transcriptase and PCR.
B) Restriction endonuclease recognition sites produce DNA fragments that differ in size when the source of the DNA is from different chromosomes; RFLPs are any sequence of RNA amplified by reverse transcriptase and PCR.
C) Restriction endonuclease recognition sites produce DNA fragments that differ in size when the source of the DNA is from different chromosomes; RFLPs are any sequence of DNA recognized by a restriction endonuclease.
D) Restriction endonuclease recognition sites are any sequence of DNA recognized by a restriction endonuclease; RFLPs are DNA fragments produced by restriction endonuclease digestion that differ in size when DNA sequences differ between chromosomes or individuals.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
30
The basic idea of using RFLPs as markers of genetic diseases is to
A) calculate the frequency of a RFLP allele and a disease phenotype.
B) determine that a particular locus gives restriction endonuclease digestion fragments of different sizes.
C) establish close linkage between a RFLP allele and a disease-associated allele.
D) generate a genetic map using biochemical markers, such as the presence or absence or enzyme activity, and correlate these markers with the presence or absence of a disease.
A) calculate the frequency of a RFLP allele and a disease phenotype.
B) determine that a particular locus gives restriction endonuclease digestion fragments of different sizes.
C) establish close linkage between a RFLP allele and a disease-associated allele.
D) generate a genetic map using biochemical markers, such as the presence or absence or enzyme activity, and correlate these markers with the presence or absence of a disease.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
31
What does it mean to say that a genetic marker and a disease gene are closely linked?
A) The genetic marker lies within the coding region for the disease gene.
B) The genetic marker and the disease gene are in close physical proximity on the same chromosome and tend to be inherited together.
C) The genetic marker and the disease gene are on different chromosomes.
D) The DNA sequence of the genetic marker and the DNA sequence of the disease gene are nearly identical.
A) The genetic marker lies within the coding region for the disease gene.
B) The genetic marker and the disease gene are in close physical proximity on the same chromosome and tend to be inherited together.
C) The genetic marker and the disease gene are on different chromosomes.
D) The DNA sequence of the genetic marker and the DNA sequence of the disease gene are nearly identical.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
32
When performing gel electrophoresis, why do molecular biologists add a "ladder" made up of nucleic acids or proteins of known size?
A) To help determine the size of each macromolecule after the separation has occurred.
B) To help macromolecules move through the gel more rapidly.
C) To make sure the dye included in the sample buffer doesn't interfere with the movement of the macromolecules that are being studied.
D) The macromolecules being studied would be invisible without the ladder.
A) To help determine the size of each macromolecule after the separation has occurred.
B) To help macromolecules move through the gel more rapidly.
C) To make sure the dye included in the sample buffer doesn't interfere with the movement of the macromolecules that are being studied.
D) The macromolecules being studied would be invisible without the ladder.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
33
Which diagram (I or II) shows the correct drawing of gel electrophoresis of DNA fragments of differing size? ? 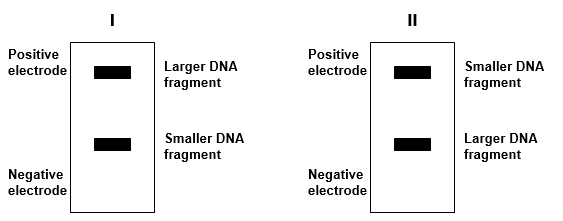
A) I
B) II

A) I
B) II

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
34
To analyze the results of a DNA sequencing reaction, which type of gel should you prepare?
A) denaturing polyacrylamide gel
B) agarose gel
C) pulsed field gel
D) nondenaturing polyacrylamide gel
A) denaturing polyacrylamide gel
B) agarose gel
C) pulsed field gel
D) nondenaturing polyacrylamide gel

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
35
Which step in Southern blotting allows an investigator to identify a particular DNA fragment that is present within a complex mixture of fragments?
A) restriction endonuclease digestion of DNA
B) agarose gel electrophoresis
C) blotting
D) hybridization with a labeled probe
A) restriction endonuclease digestion of DNA
B) agarose gel electrophoresis
C) blotting
D) hybridization with a labeled probe

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
36
Pedigree analysis was used to determine whether a genetic marker is inherited with a disease trait (black circle). Results of Southern blot analysis of restriction endonuclease-digested DNA from members of the family are shown. What conclusions can be drawn from the data? 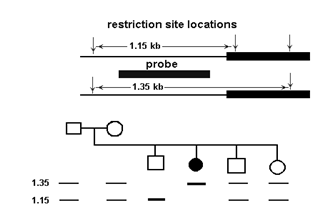
A) A restriction fragment length polymorphism (RFLP) is linked to this autosomal recessivetrait.
B) A restriction fragment length polymorphism (RFLP) is linked to this autosomal dominanttrait.
C) The disease is caused by expansion of a trinucleotide repeat.
D) No information can be gained from this analysis because researchers should haveperformed a Northern blot, not a Southern blot.

A) A restriction fragment length polymorphism (RFLP) is linked to this autosomal recessivetrait.
B) A restriction fragment length polymorphism (RFLP) is linked to this autosomal dominanttrait.
C) The disease is caused by expansion of a trinucleotide repeat.
D) No information can be gained from this analysis because researchers should haveperformed a Northern blot, not a Southern blot.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
37
Pedigree analysis was used to determine whether a genetic marker is inherited with a disease trait (black symbols). Results of Southern blot analysis of restriction endonuclease-digested DNA from members of the family are shown. What conclusions can be drawn from the data? 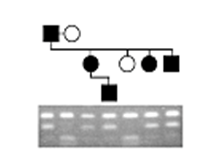
A) A restriction fragment length polymorphism (RFLP) is linked to this autosomal recessivetrait.
B) A restriction fragment length polymorphism (RFLP) is linked to this autosomal dominanttrait.
C) The disease is caused by expansion of a trinucleotide repeat.
D) No information can be gained from this analysis because researchers should haveperformed a Northern blot, not a Southern blot.

A) A restriction fragment length polymorphism (RFLP) is linked to this autosomal recessivetrait.
B) A restriction fragment length polymorphism (RFLP) is linked to this autosomal dominanttrait.
C) The disease is caused by expansion of a trinucleotide repeat.
D) No information can be gained from this analysis because researchers should haveperformed a Northern blot, not a Southern blot.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
38
Dideoxy DNA sequencing is based on chain termination with dideoxynucleotides. Why are normal deoxynucleotides also included in the reaction?
A) to act as a primer for chain termination by the dideoxynucleotides
B) to provide a template for DNA polymerase
C) to allow production of a range of synthesis products of different lengths.
D) to provide a 2?-OH required for incorporation of the dideoxynucleotides
A) to act as a primer for chain termination by the dideoxynucleotides
B) to provide a template for DNA polymerase
C) to allow production of a range of synthesis products of different lengths.
D) to provide a 2?-OH required for incorporation of the dideoxynucleotides

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
39
It is common to use ddNTPs (dideoxynucleoside triphosphates) for sequencing DNA because, when incorporated into a growing DNA polymer, the ddNTP acts as a chain terminator, allowing the researcher an opportunity to determine the last-added base to a given chain. From your knowledge of nucleotide structure and DNA synthesis, what is the most likely attribute of ddNTPs for DNA sequencing?
A) The ddNTPs lack a nitrogenous base.
B) The ddNTPs lack the sugar component.
C) The ddNTPs lack a 5 ' carbon.
D) The ddNTPs lack a 3 ' hydroxyl group.
A) The ddNTPs lack a nitrogenous base.
B) The ddNTPs lack the sugar component.
C) The ddNTPs lack a 5 ' carbon.
D) The ddNTPs lack a 3 ' hydroxyl group.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
40
The smallest fragment at the end of a DNA sequencing gel represents the
A) 2? end of the newly synthesized DNA.
B) 3? end of the newly synthesized DNA.
C) 5? end of the newly synthesized DNA.
D) either end of newly synthesized DNA, depending on how the sample was loaded on the gel.
A) 2? end of the newly synthesized DNA.
B) 3? end of the newly synthesized DNA.
C) 5? end of the newly synthesized DNA.
D) either end of newly synthesized DNA, depending on how the sample was loaded on the gel.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
41
The sequence read from a DNA sequencing gel is
A) the same sequence as the original strand.
B) complementary to the original strand.
C) the same polarity as the original strand.
D) both B and C
A) the same sequence as the original strand.
B) complementary to the original strand.
C) the same polarity as the original strand.
D) both B and C

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
42
Automated DNA sequencing differs from manual DNA sequence in which way?
A) Instead of having to run four separate sequencing reactions, in automated sequencing thereactions can be combined into one tube.
B) ddNTPs are not used in automated DNA sequencing.
C) Automated DNA sequencing uses radioactive markers.
D) In automated DNA sequencing, DNA fragments are separated by sequence instead of bysize.
A) Instead of having to run four separate sequencing reactions, in automated sequencing thereactions can be combined into one tube.
B) ddNTPs are not used in automated DNA sequencing.
C) Automated DNA sequencing uses radioactive markers.
D) In automated DNA sequencing, DNA fragments are separated by sequence instead of bysize.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
43
Which of the following statements regarding 454 pyrosequencing is not true?
A) Each DNA fragment to be sequenced must be clonally amplified.
B) Dideoxy nucleotides are used to reversibly terminate chain elongation.
C) Nucleotide incorporation is detected as light generated by the luciferase enzyme.
D) Many sequencing reactions are carried in in parallel in a multi-well plate.
A) Each DNA fragment to be sequenced must be clonally amplified.
B) Dideoxy nucleotides are used to reversibly terminate chain elongation.
C) Nucleotide incorporation is detected as light generated by the luciferase enzyme.
D) Many sequencing reactions are carried in in parallel in a multi-well plate.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
44
Pulse-field electrophoresis is useful for separating
A) small single-stranded RNAs
B) very large DNA molecules
C) degenerate oligonucleotide probes
D) DNA-RNA hybrid duplexes
A) small single-stranded RNAs
B) very large DNA molecules
C) degenerate oligonucleotide probes
D) DNA-RNA hybrid duplexes

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
45
To read an old-fashioned Sanger sequencing gel in the 5' to 3' direction, on must read the gel
A) from top to bottom.
B) from bottom to top.
C) from left to right.
D) from right to left.
A) from top to bottom.
B) from bottom to top.
C) from left to right.
D) from right to left.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
46
The polymerase chain reaction does not require
A) dNTPs
B) DNA ligase
C) DNA polymerase
D) primers
A) dNTPs
B) DNA ligase
C) DNA polymerase
D) primers

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
47
Define the term "restriction endonuclease" and explain the function of restriction endonucleases in bacteria.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
48
Describe some of the key experiments and discoveries leading to the development of recombinant DNA technology and the first cloning experiments.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
49
When restriction endonucleases first bind to DNA they random walk, slide, hop, and jump. Why don't they immediately get on with their business of cutting DNA?

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
50
Why is sulfur-35 (35S) used to label proteins but not nucleic acids?

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
51
Explain the terms "random walk" and "coupling" in the context of the mode of action of restriction endonucleases.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
52
Describe X-ray crystallographic evidence for the mode of action of EcoRI.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
53
You want to join together two pieces of DNA that have been cut with SmaI. Outline a strategy you could use to increase the efficiency of blunt end ligation.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
54
Why do foreign DNAs need to be inserted into vectors to clone them?

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
55
You want to clone a 1.5 kb cDNA. Which vectors discussed would be appropriate to use? Which would be inappropriate? Explain your answer.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
56
You want to make a genomic library with DNA fragments averaging about 85 kb in length. Which vectors discussed would be appropriate to use? Which would be inappropriate? Explain your answer.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
57
Describe the process of cloning a DNA fragment into the multiple cloning site of the vector pUC18. How would you screen for clones that contain an insert?

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
58
Explain the underlying principle between blue-white colony screening.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
59
Describe the process of cloning a DNA fragment into bacteriophage lambda. How would you screen for clones that contain an insert?

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
60
Describe the process of cloning a DNA fragment into a YAC. How would you screen for clones that contain an insert?

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
61
Compare and contrast genomic libraries with cDNA libraries.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
62
Describe the principle of ion-exchange chromatography. Describe the behavior of a positively charged protein and a negatively charged protein in a column containing positively charged resin during sample loading and elution.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
63
Describe the principle of gel filtration chromatography. For two proteins of different size, illustrate in which fraction they will elute.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
64
Describe the principle of antibody-affinity chromatography. For one protein that is recognized by the antibody and a second protein that is not, illustrate at which point each protein will be released from the column.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
65
Diagram the process for creating a double-stranded cDNA.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
66
Explain why DNA polymerase from a thermophilic bacterium is used in PCR instead of DNA polymerase I from E. coli.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
67
Compare and contrast the principles of autoradiography and phosphorimaging.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
68
What are radioisotopes and why are they useful in molecular biology research?

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
69
Define the term "probe" and describe two important characteristics of a probe.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
70
What are the relative advantages and disadvantages of end-labeling versus internal-labeling a probe?

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
71
(a) Outline the steps that would be necessary to generate a cDNA library from all of the genes expressed by adenocarcinoma cells of a breast tumor. (b) How could tumor-specific transcripts be identified from this library? (Hint: think about how you could use non-cancerous breast epithelial cells as a comparison.)

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
72
Compare and contrast degenerate probes and EST-based probes.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
73
Diagram a strategy for screening a cDNA library.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
74
If you had an antibody to the protein encoded by an unknown gene of interest, what type of library could you screen to identify the sequence of the gene? Using a diagram, explain how this would work.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
75
Show how to use restriction mapping to determine the orientation of a restriction fragment ligated into a restriction site in a vector.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
76
Diagram the process of Southern blotting and probing to detect a DNA of interest. What kinds of information can be obtained from a Southern blot?

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
77
Compare and contrast the key characteristics of automated sequencing using the Sanger method and 454 pyrosequencing.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
78
You have attempted to ligate a 1.5 kb fragment of foreign DNA into the EcoRI site in the multiple cloning site of the 4.0 kb plasmid vector shown below:
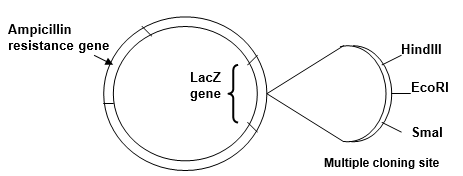 (a) After ligation you use the DNA in the ligation mixture to transform host bacteria. Why is it important to use host bacteria that are deficient for restriction-modification?
(a) After ligation you use the DNA in the ligation mixture to transform host bacteria. Why is it important to use host bacteria that are deficient for restriction-modification?
(b) You screen the bacteria that supposedly have been transformed with recombinant plasmid DNA. Some of the bacterial colonies growing on the nutrient agar plate that contains ampicillin and X-gal are white and some are blue. Explain these results.
(c) To confirm the presence of the foreign DNA insert, you perform EcoRI restriction endonuclease digests on DNA extracted from bacterial colonies. On the diagram of an agarose gel shown below, draw the pattern of bands (label their size in kb) you would expect to see for EcoRI-digested recombinant plasmid and EcoRI-digested non-recombinant plasmid vector, after electrophoresis and staining of the gel with ethidium bromide.
 (a) After ligation you use the DNA in the ligation mixture to transform host bacteria. Why is it important to use host bacteria that are deficient for restriction-modification?
(a) After ligation you use the DNA in the ligation mixture to transform host bacteria. Why is it important to use host bacteria that are deficient for restriction-modification?(b) You screen the bacteria that supposedly have been transformed with recombinant plasmid DNA. Some of the bacterial colonies growing on the nutrient agar plate that contains ampicillin and X-gal are white and some are blue. Explain these results.
(c) To confirm the presence of the foreign DNA insert, you perform EcoRI restriction endonuclease digests on DNA extracted from bacterial colonies. On the diagram of an agarose gel shown below, draw the pattern of bands (label their size in kb) you would expect to see for EcoRI-digested recombinant plasmid and EcoRI-digested non-recombinant plasmid vector, after electrophoresis and staining of the gel with ethidium bromide.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
79
A chromotography column in which oligo-dT is linked to an inert substance is useful in separating eukaryotic mRNA from other RNA molecules. On what principle does this column operate?

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck
80
Starting with the nucleotide sequence of the human DNA ligase I gene, describe how you would search for a homologous gene in another organism whose genome has been sequenced, such as the pufferfish Tetraodon nigroviridis. Then, describe how you would obtain the protein and test it for ligase activity.

Unlock Deck
Unlock for access to all 85 flashcards in this deck.
Unlock Deck
k this deck


