Deck 10: Microbial Genomics
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/66
Play
Full screen (f)
Deck 10: Microbial Genomics
1
The Illumina sequencing method is not completely described.Given what you read about it in the text,and given that the method relies upon sequential additions of a single nucleotide,where might you expect the fluorescent removable tag to reside?
A)on a 3' OH-blocker
B)on a 5' phosphate-group
C)on a 2' OH-blocker
D)on the organic base
E)none is a logical position
A)on a 3' OH-blocker
B)on a 5' phosphate-group
C)on a 2' OH-blocker
D)on the organic base
E)none is a logical position
A
2
Sequence reading accuracy is improved with the combined use of:
A)the longer error-prone reads generated by Illumina sequencing and the shorter accuate reads generated by PacBio technology
B)the longer error-prone reads generated by PacBio sequencing and the shorter accuate reads generated by Illumina technology
C)the longer error-prone reads generated by Ion torrent sequencing and the shorter accuate reads generated by PacBio technology
D)the longer error-prone reads generated by Ion torrent sequencing and the shorter accuate reads generated by Illumina technology
E)the longer error-prone reads generated by Illumina sequencing and the shorter accuate reads generated by Ion torrent technology
A)the longer error-prone reads generated by Illumina sequencing and the shorter accuate reads generated by PacBio technology
B)the longer error-prone reads generated by PacBio sequencing and the shorter accuate reads generated by Illumina technology
C)the longer error-prone reads generated by Ion torrent sequencing and the shorter accuate reads generated by PacBio technology
D)the longer error-prone reads generated by Ion torrent sequencing and the shorter accuate reads generated by Illumina technology
E)the longer error-prone reads generated by Illumina sequencing and the shorter accuate reads generated by Ion torrent technology
B
3
In pyrosequencing,which enzyme creates ATP?
A)DNA polymerase
B)Luciferase
C)ATP sulfurylase
D)Restriction endonuclease
E)DNA ligase
A)DNA polymerase
B)Luciferase
C)ATP sulfurylase
D)Restriction endonuclease
E)DNA ligase
C
4
What might be a familiar term to use in instead of "PCR adapters" as we examine the 454 pyrosequencing technique?
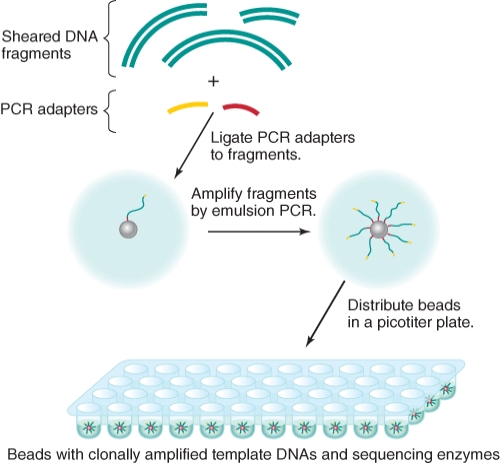
A)multiple cloning sites
B)restriction enzyme sites
C)generic primer recognition sites
D)DNA polymerase recognition sites
E)DNA ligase sites

A)multiple cloning sites
B)restriction enzyme sites
C)generic primer recognition sites
D)DNA polymerase recognition sites
E)DNA ligase sites

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
5
Many scientists deposit their gene sequences in GenBank to make them publicly available to other scientist via the internet.Approximately how many base pairs have been deposited thus far?
A)10,000
B)100,000
C)1 million
D))10 million
E)greater than 10 million
A)10,000
B)100,000
C)1 million
D))10 million
E)greater than 10 million

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
6
454 pyrosequencing coupled advances in:
A)DNA sequencing and DNA amplification
B)DNA sequencing and DNA cloning
C)DNA cloning and DNA amplification
D)Radiolabeling and DNA cloning
E)Fluorescence labeling and DNA cloning
A)DNA sequencing and DNA amplification
B)DNA sequencing and DNA cloning
C)DNA cloning and DNA amplification
D)Radiolabeling and DNA cloning
E)Fluorescence labeling and DNA cloning

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
7
Why is it difficult to define "species" accurately when considering bacterial life forms? (Select all that apply)
A)genome sizes vary considerable
B)novel genes are found in some species members
C)bacteria do not reproduce sexually
D)mutation is ongoing and complicates sequence comparisons
E)all are correct choices
A)genome sizes vary considerable
B)novel genes are found in some species members
C)bacteria do not reproduce sexually
D)mutation is ongoing and complicates sequence comparisons
E)all are correct choices

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
8
The "paired-end read" approach of Illumina sequencing suggests an approach in constructing DNA fragments to be read which is similar to that used in :
A)Sanger dideoxy
B)Pyrosequencing
C)Ion torrent sequencing
D)Maxam and gilbertsequencing
E)None of the approaches generate non-cloned DNA fragments readable in both directions
A)Sanger dideoxy
B)Pyrosequencing
C)Ion torrent sequencing
D)Maxam and gilbertsequencing
E)None of the approaches generate non-cloned DNA fragments readable in both directions

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
9
Which of these is another term for the Sanger method of DNA sequencing?
A)chemical degradation method
B)dideoxy sequencing.
C)restriction method
D)label method
E)gel method
A)chemical degradation method
B)dideoxy sequencing.
C)restriction method
D)label method
E)gel method

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
10
In pyrosequencing,which enzyme generates the light detected by the CCD device?
A)DNA polymerase
B)Luciferase
C)ATP sulfurylase
D)Restriction endonuclease
E)DNA ligase
A)DNA polymerase
B)Luciferase
C)ATP sulfurylase
D)Restriction endonuclease
E)DNA ligase

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
11
The Sanger method of DNA sequencing has been automated and uses a(n):
A)thermal-stable DNA polymerase to allow for multiple rounds of DNA synthesis.
B)trideoxynucleotides in the synthesis reactions.
C)none is a correct choice
D)agarose gel electrophoresis for fragment separation.
E)chemical digestion method for fragment generation.
A)thermal-stable DNA polymerase to allow for multiple rounds of DNA synthesis.
B)trideoxynucleotides in the synthesis reactions.
C)none is a correct choice
D)agarose gel electrophoresis for fragment separation.
E)chemical digestion method for fragment generation.

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
12
What is a transcriptome?
A)The coding region on the chromosome.
B)The coding region on the chromosome minus the introns.
C)The coding region on the chromosome plus the control regions.
D)The transcripts encoded for by the genes within a genome.
E)The mount of mRNA in the cell.
A)The coding region on the chromosome.
B)The coding region on the chromosome minus the introns.
C)The coding region on the chromosome plus the control regions.
D)The transcripts encoded for by the genes within a genome.
E)The mount of mRNA in the cell.

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
13
The technical term to describe apyrase activity is:
A)nucleotidase
B)nuclease
C)exonuclease
D)endonuclease
E)nucleosidase
A)nucleotidase
B)nuclease
C)exonuclease
D)endonuclease
E)nucleosidase

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
14
The sequencing method which uses detection of pyrophosphate liberated when nucleotides are added to a growing DNA chain is called:
A)Sanger dideoxy sequencing
B)Plus-minus sequencing
C)Pyrosequencing
D)Ion torrent sequencing
E)PacBio sequencing
A)Sanger dideoxy sequencing
B)Plus-minus sequencing
C)Pyrosequencing
D)Ion torrent sequencing
E)PacBio sequencing

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
15
Adenosine phosphosulfate is a chemical analog of
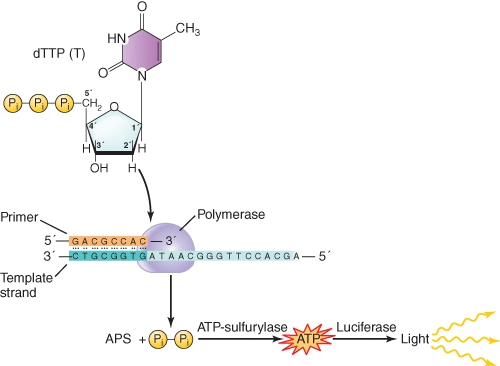
A)ADP
B)ATP
C)AMP
D)Pyrophosphate
E)Phosphate

A)ADP
B)ATP
C)AMP
D)Pyrophosphate
E)Phosphate

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
16
In pyrosequencing,which enzyme liberates pyrophosphate?
A)DNA polymerase
B)Luciferase
C)ATP sulfurylase
D)Restriction endonuclease
E)DNA ligase
A)DNA polymerase
B)Luciferase
C)ATP sulfurylase
D)Restriction endonuclease
E)DNA ligase

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
17
The Sanger method of sequencing uses ALL of the following EXCEPT:
A)dideoxynucleotides.
B)DNA polymerase.
C)a short oligo primer.
D)deoxynucleotides
E)restriction enzymes.
A)dideoxynucleotides.
B)DNA polymerase.
C)a short oligo primer.
D)deoxynucleotides
E)restriction enzymes.

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
18
Ion torrent sequencing detects:
A)electrons liberated as nucleotides are incorporated into DNA
B)protons liberated as nucleotides are incorporated into DNA
C)pyrophosphate liberated as nucleotides are incorporated into DNA
D)the nucleotide found at the 3' end of a DNA fragment
E)the nucleotide found at the 5' end of a DNA fragment
A)electrons liberated as nucleotides are incorporated into DNA
B)protons liberated as nucleotides are incorporated into DNA
C)pyrophosphate liberated as nucleotides are incorporated into DNA
D)the nucleotide found at the 3' end of a DNA fragment
E)the nucleotide found at the 5' end of a DNA fragment

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
19
What can you infer from reading the description of 454 pyrosequencing?
A)sheared DNA initially generated very short DNA fragments
B)sheared DNA initially generated very long DNA fragments
C)dilution of DNA to be sequenced presented initial challenges
D)PCR optimization was challenging in developing this techniique
E)None is a plausible inference
A)sheared DNA initially generated very short DNA fragments
B)sheared DNA initially generated very long DNA fragments
C)dilution of DNA to be sequenced presented initial challenges
D)PCR optimization was challenging in developing this techniique
E)None is a plausible inference

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
20
What is an open-reading frame?
A)A non-translated region on the chromosome.
B)an unknown gene.
C)A gene that no longer has a function.
D)The protein encoding sequence of a gene.
E)The intron region of a gene.
A)A non-translated region on the chromosome.
B)an unknown gene.
C)A gene that no longer has a function.
D)The protein encoding sequence of a gene.
E)The intron region of a gene.

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
21
A bacterial genome has an overall GC content of 60%.Two percent of the genome is considered to be a symbiosis island whose GC content is 40%.What GC richness does the rest of the genome actually possess?
A)70%
B)65%
C)60%
D)55%
E)85%
A)70%
B)65%
C)60%
D)55%
E)85%

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
22
Which of these can be used to determine protein structure?
A)amino acid sequencing
B)X-ray crystallography and NMR
C)polyacrylamide gel electrophoresis (PAGE)
D)denaturing gel electrophoresis
E)mass spectrometry
A)amino acid sequencing
B)X-ray crystallography and NMR
C)polyacrylamide gel electrophoresis (PAGE)
D)denaturing gel electrophoresis
E)mass spectrometry

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
23
Genes which confer invasiveness to a pathogen and whose GC content varies significantly from that of the totla genome constitute;
A)Paralogs
B)Orthologs
C)Pathogenicity islands
D)Symbiosis islands
E)SSU rRNA genes
A)Paralogs
B)Orthologs
C)Pathogenicity islands
D)Symbiosis islands
E)SSU rRNA genes

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
24
Term for genes from different organisms that encode for proteins that carry out the same function but have different sequences.
A)homologs
B)paralogs
C)orthologs
D)metalogs
E)semilogs
A)homologs
B)paralogs
C)orthologs
D)metalogs
E)semilogs

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
25
Two-dimensional polyacrylamide gel electrophoresis (2D-PAGE)separates proteins based on what property?
A)size
B)charge
C)hydrophobicity
D)size and charge
E)size and hydrophobicity
A)size
B)charge
C)hydrophobicity
D)size and charge
E)size and hydrophobicity

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
26
Suppose it is observed that a small section of a microorganism's chromosome varies significantly in GC content from the majority of the chromosome.What likely caused this?
A)spontaneous mutations
B)induced mutations
C)horizontal gene transfer
D)vertical gene transfer
E)site-directed mutagenesis
A)spontaneous mutations
B)induced mutations
C)horizontal gene transfer
D)vertical gene transfer
E)site-directed mutagenesis

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
27
What is the isoelectric point of a protein?
A)The pH at which the protein has the greatest charge.
B)The pH at which a protein has a negative charge.
C)The pH at which a protein has no charge.
D)The proton potential of a protein in an acrylamide gel.
E)The proton potential of a protein in a cell.
A)The pH at which the protein has the greatest charge.
B)The pH at which a protein has a negative charge.
C)The pH at which a protein has no charge.
D)The proton potential of a protein in an acrylamide gel.
E)The proton potential of a protein in a cell.

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
28
What is a proteome?
A)The coding region on the chromosome.
B)The coding region on the chromosome minus the introns.
C)The coding region on the chromosome plus the control regions.
D)The translational products encoded for by the genes within a genome.
E)The mount of mRNA in the cell.
A)The coding region on the chromosome.
B)The coding region on the chromosome minus the introns.
C)The coding region on the chromosome plus the control regions.
D)The translational products encoded for by the genes within a genome.
E)The mount of mRNA in the cell.

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
29
In order to associate a DNA gene sequence obtained through metagenomic analysis with an organism,the most reliable gene to find associated with it is:
A)tRNA gene
B)DNA polymerase
C)Dehydrogenase gene
D)SSU rRNA gene
E)Cytochrome gene
A)tRNA gene
B)DNA polymerase
C)Dehydrogenase gene
D)SSU rRNA gene
E)Cytochrome gene

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
30
What was the Northern blot hybridization technique used to measure?
A)The transcriptional expression of a specific gene.
B)The translational expression of a specific gene.
C)The frequency of transcription initiation.
D)The amount of DNA in a cell.
E)The rate of chromosomal replication.
A)The transcriptional expression of a specific gene.
B)The translational expression of a specific gene.
C)The frequency of transcription initiation.
D)The amount of DNA in a cell.
E)The rate of chromosomal replication.

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
31
A bacterial genome has an overall GC content of 60%.Ten percent of the genome is considered to be a virulence island whose GC content is 50%.What GC richness does the rest of the genome actually possess?
A)70%
B)65%
C)61%
D)55%
E)85%
A)70%
B)65%
C)61%
D)55%
E)85%

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
32
RNA-seq technology is used in preference to DNA microarray technology primarily because of :
A)RNA stability issues
B)Better accuracy in quantifying gene expression levels
C)Better detection technology
D)More affordability
E)None is the correct reason
A)RNA stability issues
B)Better accuracy in quantifying gene expression levels
C)Better detection technology
D)More affordability
E)None is the correct reason

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
33
Which of these is TRUE regarding horizontal gene transfer?
A)It does not result in permanent change.
B)It does not play a role in microbial evolution.
C)It does not result in any significant consequences for the organism involved.
D)It is always occurring in nature.
E)It does not alter the genome of the organism.
A)It does not result in permanent change.
B)It does not play a role in microbial evolution.
C)It does not result in any significant consequences for the organism involved.
D)It is always occurring in nature.
E)It does not alter the genome of the organism.

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
34
What are paralogs?
A)Genes that arise from a duplication event in an organism.
B)Mutant genes in an organism.
C)Genes that are seldom expressed in an organism.
D)Genes that are expressed at a high level in an organism.
E)Genes that have no function in an organism.
A)Genes that arise from a duplication event in an organism.
B)Mutant genes in an organism.
C)Genes that are seldom expressed in an organism.
D)Genes that are expressed at a high level in an organism.
E)Genes that have no function in an organism.

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
35
If microbes are not capable of growth as pure isolated cultures,which factor do you consider the most likely to contribute to the growth challenge?
A)Unculturable microbes may receive essential nutrients as a result of the metabolic activity of another member of a mixed microbial community
B)Nutrients not yet identified by bioscientists may be lacking from a defined growth medium
C)Gene transfer from other microbes may be required for active growth
D)RNA transfer from other microbes may be required for active growth
E)Protein transfer from other microbes may be required for active growth
A)Unculturable microbes may receive essential nutrients as a result of the metabolic activity of another member of a mixed microbial community
B)Nutrients not yet identified by bioscientists may be lacking from a defined growth medium
C)Gene transfer from other microbes may be required for active growth
D)RNA transfer from other microbes may be required for active growth
E)Protein transfer from other microbes may be required for active growth

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
36
DNA microarray analysis may be used to study:
A)gene expression in cells grown under different conditions.
B)protein expression in cells grown under different conditions.
C)rate of chromosomal replication in cells.
D)enzyme activity in cells.
E)growth rate of cells when grown under different conditions.
A)gene expression in cells grown under different conditions.
B)protein expression in cells grown under different conditions.
C)rate of chromosomal replication in cells.
D)enzyme activity in cells.
E)growth rate of cells when grown under different conditions.

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
37
In RNA-seq technology,it is stated that rRNA is discarded before analysis.Why do you think this is done?
A)rRNA will be abundant compared with other transcripts,potentially making their expression difficult to detect
B)rRNA binds to other transcripts and degrades them
C)rRNA cannot be converted to cDNA
D)rRNA is less stable than other RNA forms and will release small interfering fragments
E)none is a plausible reason
A)rRNA will be abundant compared with other transcripts,potentially making their expression difficult to detect
B)rRNA binds to other transcripts and degrades them
C)rRNA cannot be converted to cDNA
D)rRNA is less stable than other RNA forms and will release small interfering fragments
E)none is a plausible reason

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
38
Which of these methods allows for recovery of single cells from a mixed microbial community?
A)RNA-seq
B)DNA microarray technology
C)Fluorescence-activated cell sorting
D)Illumina technology
E)All technologies are useful
A)RNA-seq
B)DNA microarray technology
C)Fluorescence-activated cell sorting
D)Illumina technology
E)All technologies are useful

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
39
Why might both restriction enzyme digestion and sonication be required for the success of a shotgun sequencing approach? (Select all that apply)
A)Too many restriction fragments might otherwise be generated
B)Too few restriction fragments might otherwise be generated
C)Further validation of nucleotide sequence order and arrangement may be obtained through comparison of sequence data derived from each method
D)All responses are correct
E)No responses are relevant
A)Too many restriction fragments might otherwise be generated
B)Too few restriction fragments might otherwise be generated
C)Further validation of nucleotide sequence order and arrangement may be obtained through comparison of sequence data derived from each method
D)All responses are correct
E)No responses are relevant

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
40
What is metaproteomics?
A)A culture-dependent method that may be used to determine protein sequences.
B)A culture-dependent method that may be used to find new enzymes.
C)A culture-dependent method that may be used to study groups of microbes in an ecosystem.
D)A culture-independent method that may be used to document microbial community enzyme pools
E)A culture -independent method that may be used for the isolation of different organisms.
A)A culture-dependent method that may be used to determine protein sequences.
B)A culture-dependent method that may be used to find new enzymes.
C)A culture-dependent method that may be used to study groups of microbes in an ecosystem.
D)A culture-independent method that may be used to document microbial community enzyme pools
E)A culture -independent method that may be used for the isolation of different organisms.

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
41
Paralogs are genes that arose from a duplication event in the cell.

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
42
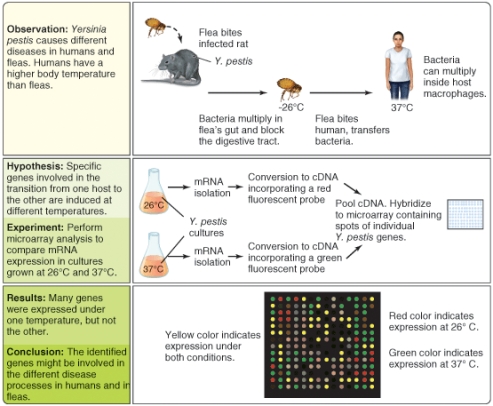
What types of gene do you think might generate green dots in the DNA microarray pattern? (Select all that apply)
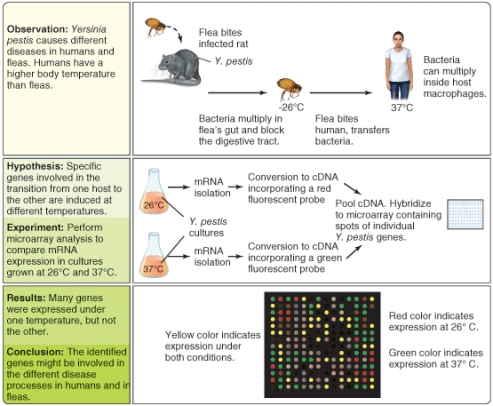
A)genes for amino acid biosynthesis
B)genes for catabism of carbohydrates
C)virulence genes for invasion of mammals
D)virulence genes for insectoid pathogenicity
E)genes for nucleic acid biosynthesis

A)genes for amino acid biosynthesis
B)genes for catabism of carbohydrates
C)virulence genes for invasion of mammals
D)virulence genes for insectoid pathogenicity
E)genes for nucleic acid biosynthesis

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
43
Techniques that have been used to analyse the proteome in cells include: (Select all that apply)
A)X-ray crystallography
B)Northern blot analysis
C)LC-MS
D)DNA microarray analysis
E)NMR
A)X-ray crystallography
B)Northern blot analysis
C)LC-MS
D)DNA microarray analysis
E)NMR

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
44
Pyrosequencing relies on which of the following enzymes? (Select all that apply)
A)DNA polymerase
B)Luciferase
C)ATP sulfurylase
D)Restriction endonuclease
E)DNA ligase
A)DNA polymerase
B)Luciferase
C)ATP sulfurylase
D)Restriction endonuclease
E)DNA ligase

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
45
The Southern blot uses a DNA probe to hybridize to DNA fragments that have been separated by electrophoresis.

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
46
What types of gene do you think might generate red dots in the DNA microarray pattern? (Select all that apply)
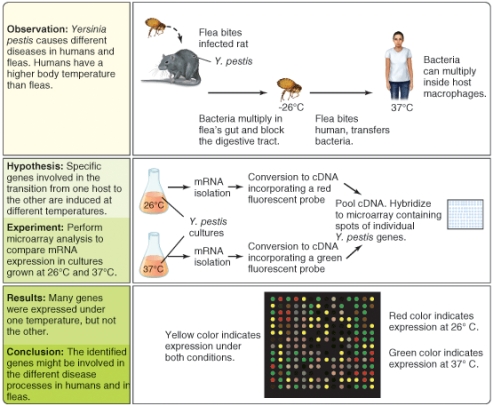
A)genes for amino acid biosynthesis
B)genes for catabism of carbohydrates
C)virulence genes for invasion of mammals
D)virulence genes for insectoid pathogenicity
E)genes for nucleic acid biosynthesis

A)genes for amino acid biosynthesis
B)genes for catabism of carbohydrates
C)virulence genes for invasion of mammals
D)virulence genes for insectoid pathogenicity
E)genes for nucleic acid biosynthesis

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
47
Metagenomics uses a culture-independent based genomic analysis of microbial communities to determine community composition.

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
48
Illumina sequencing relies upon: (Select all that apply)
A)removal of fluorescent labels on nucleotides
B)addition of fluorescently labeld nucleotides
C)DNA polymerase activity
D)DNA ligase activity
E)CCD camera detection
A)removal of fluorescent labels on nucleotides
B)addition of fluorescently labeld nucleotides
C)DNA polymerase activity
D)DNA ligase activity
E)CCD camera detection

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
49
What types of gene do you think might generate yellow dots in the DNA microarray pattern? (Select all that apply)
A)genes for amino acid biosynthesis
B)genes for catabism of carbohydrates
C)virulence genes for invasion of mammals
D)virulence genes for insectoid pathogenicity
E)genes for nucleic acid biosynthesis

A)genes for amino acid biosynthesis
B)genes for catabism of carbohydrates
C)virulence genes for invasion of mammals
D)virulence genes for insectoid pathogenicity
E)genes for nucleic acid biosynthesis

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
50
To determine that a gene in one microbe is an ortholog of a gene in a second microbe,its GC content should be: (Select all that apply)
A)Similar to that of the other gene
B)Distinct from that of the gene with related function
C)Similar to that of its own SSU rRNA gene
D)Distinct from that of its own SSU rRNA gene
E)None is a relevant choice
A)Similar to that of the other gene
B)Distinct from that of the gene with related function
C)Similar to that of its own SSU rRNA gene
D)Distinct from that of its own SSU rRNA gene
E)None is a relevant choice

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
51
The Sanger method of sequencing used special nucleotides that are called ________ nucleotides.

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
52
Horizontal gene transfer occurs when an organism acquires genetic information from another organism in its own generation.

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
53
Open reading frame detection would logically recognition of: (Select all that apply)
A)DNA polymerase binding sites
B)Promoter sequences
C)Shine dalgarno sequences
D)Terminator sequences
E)Stop codon sequences
A)DNA polymerase binding sites
B)Promoter sequences
C)Shine dalgarno sequences
D)Terminator sequences
E)Stop codon sequences

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
54
To determine that a gene is a paralog of another,its GC content should be: (Select all that apply)
A)Similar to that of the gene with a related function
B)Distinct from that of the gene with related function
C)Similar to that of the SSU rRNA gene
D)Distinct from that of the SSU rRNA gene
E)None is a relevant choice
A)Similar to that of the gene with a related function
B)Distinct from that of the gene with related function
C)Similar to that of the SSU rRNA gene
D)Distinct from that of the SSU rRNA gene
E)None is a relevant choice

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
55
Analysis of the genome sequence of an organism shows that an area of the genome contains stretch of several thousand base pairs that differ significantly in % G + C content from the rest of the genome.Most likely this DNA was originally obtained from a gene duplication event.

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
56
To determine that a gene in one microbe is an ortholog of a gene in a second microbe,its GC content should be: (Select all that apply)
A)Similar to that of the other gene
B)Distinct from that of the other gene
C)Distinct from that of the other microbe's SSU rRNA gene
D)Similar to that of the other microbe's SSU rRNA gene
E)None is a relevant choice
A)Similar to that of the other gene
B)Distinct from that of the other gene
C)Distinct from that of the other microbe's SSU rRNA gene
D)Similar to that of the other microbe's SSU rRNA gene
E)None is a relevant choice

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
57
Techniques that have been used study the transcriptome in cells include: (Select all that apply)
A)LC-MS
B)Northern blot analysis
C)X-ray crystallography
D)DNA microarray analysis
E)NMR
A)LC-MS
B)Northern blot analysis
C)X-ray crystallography
D)DNA microarray analysis
E)NMR

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
58
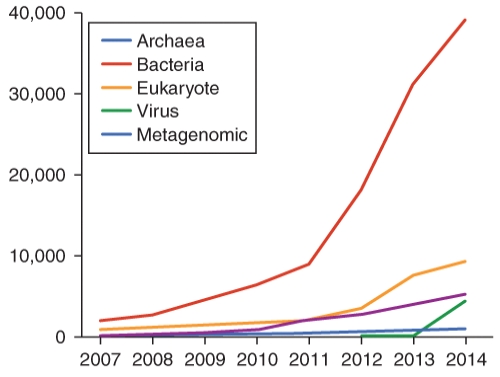
Why is is likely that bacterial DNA sequence analysis outstrips that of the other genome types? (Select all logical possibilities)
A)bacterial genomes are the smallest
B)more bacterial species are culturable than for eukaryal or archaeal species
C)bacterial genome size is relatively small and amenable to rapid sequencing
D)fewer bacterial species have been identified
E)bacterial DNA costs less per base to sequence

A)bacterial genomes are the smallest
B)more bacterial species are culturable than for eukaryal or archaeal species
C)bacterial genome size is relatively small and amenable to rapid sequencing
D)fewer bacterial species have been identified
E)bacterial DNA costs less per base to sequence

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
59
DNA microarray technology allows a researcher to analyze gene expression in an organism cultured under different growth conditions.

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
60
Techniques that have been used to analyse mRNA expression in cells include: (Select all that apply)
A)Southern blot analysis
B)Northern blot analysis
C)Western blot analysis
D)DNA microarray analysis
E)NMR
A)Southern blot analysis
B)Northern blot analysis
C)Western blot analysis
D)DNA microarray analysis
E)NMR

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
61
A culture-independent method that is used to study genomes from microbial communities in their natural environments is called _______________ .

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
62
Determining DNA sequence length through the analysis of overlapping DNA fragments is known as __________________ sequencing.

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
63
The computational techniques used to complile and analyze information generated through DNA sequencing belong to the field of science known as ___________________.

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
64
The complete complement of proteins in the cell that results from the translation of all the mRNA transcripts is called the ____________.

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
65
The pH at which a protein's charge is completely neutral is the protein's ______________ point.

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck
66
In analyzing a genome sequence,a computer program identifies the beginning and the end of a protein encoding sequence.This sequence is referred to as a(n)_______ _______ frame.

Unlock Deck
Unlock for access to all 66 flashcards in this deck.
Unlock Deck
k this deck


