Deck 16: Gene Regulation in Eukaryotes II: Epigenetics and Regulation at the RNA Level
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/36
Play
Full screen (f)
Deck 16: Gene Regulation in Eukaryotes II: Epigenetics and Regulation at the RNA Level
1
Who originally identified a highly condensed structure in the interphase of nuclei?
A)Lyon
B)Barr
C)Ohno
D)None of the answers are correct
A)Lyon
B)Barr
C)Ohno
D)None of the answers are correct
B
2
Most imprinted genes are silenced. What is one exception to this rule?
A)H19
B)Igf-2
C)UBE3A
D)SNRPN
A)H19
B)Igf-2
C)UBE3A
D)SNRPN
B
3
The expression of a single allele is associated with which of the following?
A)X-inactivation
B)Genomic imprinting
C)Maternal inheritance
D)Extra nuclear inheritance
A)X-inactivation
B)Genomic imprinting
C)Maternal inheritance
D)Extra nuclear inheritance
B
4
Which of the following statements are correct?
A)Changes in gene expression based on environmental conditions are not considered normal while developmental changes are
B)Environmental epigenetic changes can vary due to the exposure of the organism to different environmental conditions, while those programmed during development are the result of stimuli generated by the organism itself
C)Environmental epigenetic gene regulation only occurs in reptiles and insects, while developmental epigenetic regulation occurs in all animals
D)Environmental epigenetic gene regulation is typically reversible while developmental epigenetic gene regulation is typically not reversible.
A)Changes in gene expression based on environmental conditions are not considered normal while developmental changes are
B)Environmental epigenetic changes can vary due to the exposure of the organism to different environmental conditions, while those programmed during development are the result of stimuli generated by the organism itself
C)Environmental epigenetic gene regulation only occurs in reptiles and insects, while developmental epigenetic regulation occurs in all animals
D)Environmental epigenetic gene regulation is typically reversible while developmental epigenetic gene regulation is typically not reversible.

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
5
What gene is most responsible for X-inactivation?
A)Xic
B)Xist
C)Tsix
D)Xce
A)Xic
B)Xist
C)Tsix
D)Xce

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
6
Which of the following is an example of epigenetic inheritance?
A) Expression of the Igf-2 gene based on methylation of the ICR and DMR regions
B) Inheritance of flower color as studied by Mendel
C) Leaf coloration based on mitochondrial inheritance
D) Snail shell twist patterns
A) Expression of the Igf-2 gene based on methylation of the ICR and DMR regions
B) Inheritance of flower color as studied by Mendel
C) Leaf coloration based on mitochondrial inheritance
D) Snail shell twist patterns

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
7
How many Barr bodies would an individual with a XXY genotype possess?
A)0
B)1
C)2
D)None of the answers are correct
A)0
B)1
C)2
D)None of the answers are correct

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
8
A modification that occurs to a nuclear gene that alters gene expression, but not permanently, is called _______ inheritance.
A)extranuclear
B)cytoplasmic
C)maternal effect
D)epigenetic
E)nuclear
A)extranuclear
B)cytoplasmic
C)maternal effect
D)epigenetic
E)nuclear

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
9
Epigenetic inheritance may occur at which of the following stages?
A)Oogenesis
B)Spermatogenesis
C)Embryogenesis
D)All of the answers are correct
A)Oogenesis
B)Spermatogenesis
C)Embryogenesis
D)All of the answers are correct

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
10
What is the molecular mechanism for imprinting a gene?
A)Acetylation
B)Nitration
C)Phosphorylation
D)Methylation
A)Acetylation
B)Nitration
C)Phosphorylation
D)Methylation

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
11
The Lyon hypothesis attempts to explain the molecular mechanism of _____.
A)X-inactivation
B)Genomic imprinting
C)Maternal inheritance
D)Extra nuclear inheritance
A)X-inactivation
B)Genomic imprinting
C)Maternal inheritance
D)Extra nuclear inheritance

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
12
What gene(s) is/are encoded in the Xic?
A)Xce
B)Xist
C)Tsix
D)Both the Tsix and Xist genes are in the Xic region
A)Xce
B)Xist
C)Tsix
D)Both the Tsix and Xist genes are in the Xic region

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
13
Expression of ______ would inhibit X-inactivation.
A)Xic
B)Xist
C)Tsix
D)All of the answers are correct
E)None of the answers are correct
A)Xic
B)Xist
C)Tsix
D)All of the answers are correct
E)None of the answers are correct

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
14
The differentially methylated region (DMR) is associated with which of the following?
A)X-inactivation
B)Genomic imprinting
C)Maternal inheritance
D)Extra nuclear inheritance
E)All of the answers are correct
A)X-inactivation
B)Genomic imprinting
C)Maternal inheritance
D)Extra nuclear inheritance
E)All of the answers are correct

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
15
In the Igf-2 allele, which chromosome is imprinted? Which is expressed?
A)Paternal, paternal
B)Paternal, maternal
C)Maternal, paternal
D)Maternal, maternal
A)Paternal, paternal
B)Paternal, maternal
C)Maternal, paternal
D)Maternal, maternal

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
16
Both parents usually imprint the same gene.

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
17
If a cell is fused with another cell, which of the following would be the best example of a trans-mechanism of epigenetic control?
A)A gene that is originally silenced in one cell but expressed in the other is silenced in the hybrid
B)A gene that is originally silenced in one cell but expressed in the other is still expressed in the hybrid.
C)The methylation pattern of the same gene from either cell is not altered
D)There are no examples that could conform to a trans-mechanism
A)A gene that is originally silenced in one cell but expressed in the other is silenced in the hybrid
B)A gene that is originally silenced in one cell but expressed in the other is still expressed in the hybrid.
C)The methylation pattern of the same gene from either cell is not altered
D)There are no examples that could conform to a trans-mechanism

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
18
What are some diseases associated with imprinting?
A)Angelman Syndrome
B)LHON
C)Alzheimer's Disease
D)All of the answers are correct
A)Angelman Syndrome
B)LHON
C)Alzheimer's Disease
D)All of the answers are correct

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
19
Which of the following are molecular mechanisms used in epigentic gene regulation?
A) DNA methylation
B) Covalent histone modification
C) Chromatin remodeling
D) All are correct
A) DNA methylation
B) Covalent histone modification
C) Chromatin remodeling
D) All are correct

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
20
What genes appear to be controlled by the Xic?
A)Xist and Tsix
B)Xist
C)TsiX
D)Xic
A)Xist and Tsix
B)Xist
C)TsiX
D)Xic

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
21
A common way of studying methylation in cells is to sequence DNA samples before and after modification with sodium bisulfite. The sodium bisulfite deaminates Cytosine residues, generating Uracil residues, therefore resulting in a change in the sequence as compared to the non-modified DNA. Sodium bisulfite does not react with 5-methylcytosine so there will be no change in the sequence of those modified bases. When tumors are sequenced to study methylation patterns and epigenetic control, what would be the best control for sequencing using this technique?
A)Non-cancerous tissue DNA from a different individual but from the same organ
B)Liver DNA from the same individual that the cancer sample is from
C)DNA from non-cancerous tissue from the same individual's organ as the cancer
D)A combination of DNA from liver, brain and muscle from the same individual that has the cancer
A)Non-cancerous tissue DNA from a different individual but from the same organ
B)Liver DNA from the same individual that the cancer sample is from
C)DNA from non-cancerous tissue from the same individual's organ as the cancer
D)A combination of DNA from liver, brain and muscle from the same individual that has the cancer

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
22
In an in vitro translation assay radioactive amino acids can be incorporated into proteins and the proteins separated from the unincorporated amino acids. Given the following data what are the conclusions that you would come to? 
A) Double stranded RNA is more efficient at inhibiting translation than antisense RNA
B) Antisense RNA is more efficient at inhibiting translation than double stranded RNA
C) No conclusion about the efficiency of translation inhibiton can be made as the positive and negative controls did not perform appropriately
D) The experiment did not work since the antisense fibronectin did not have an effect on amino acid incorporation.

A) Double stranded RNA is more efficient at inhibiting translation than antisense RNA
B) Antisense RNA is more efficient at inhibiting translation than double stranded RNA
C) No conclusion about the efficiency of translation inhibiton can be made as the positive and negative controls did not perform appropriately
D) The experiment did not work since the antisense fibronectin did not have an effect on amino acid incorporation.

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
23
It is possible that the effects of the association of the ICR and DMR regions with the CTC factors could be distance dependent. An experimental outline could be devised where the effects of the distances of these regions could be varied and the rates of transcription could be measured. Below is a graph of some possible results. Which letter represents the most likely outcome if an increase in distance between these two regions results in a diminished effect on transcription rates? 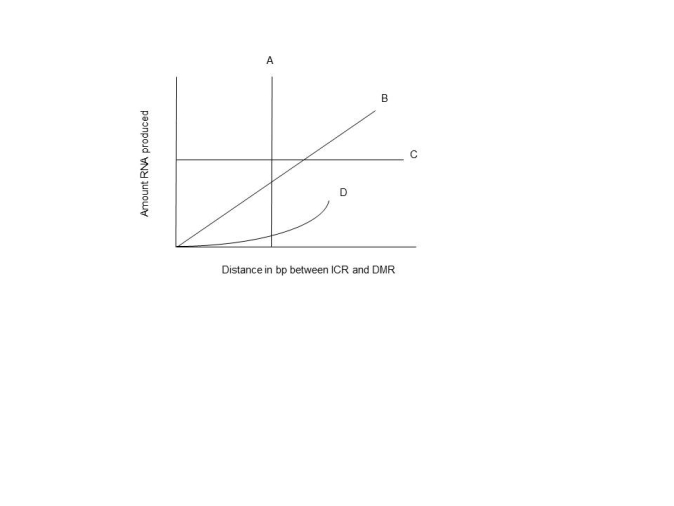
A) A
B) B
C) C
D) D

A) A
B) B
C) C
D) D

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
24
The most likely explanation for the phenomenon where a protein may be found in different sub-cellular locations in different cells is
A) stabilization of the mRNA transcript
B) alternative splicing
C) RNA interference
D) transport of mRNA from the nucleus to the cytoplasm
A) stabilization of the mRNA transcript
B) alternative splicing
C) RNA interference
D) transport of mRNA from the nucleus to the cytoplasm

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
25
What would be the consequence if an individual was homozygous for an IBP that could not bind iron but could bind to the IRE?
A) Unregulated transferrin receptor levels
B) Increased degradation of ferritin
C) Possible iron toxicity to the cells at levels of iron that would not normally be toxic to a cell
D) There would be no predicted consequence to iron metabolism in the cell
A) Unregulated transferrin receptor levels
B) Increased degradation of ferritin
C) Possible iron toxicity to the cells at levels of iron that would not normally be toxic to a cell
D) There would be no predicted consequence to iron metabolism in the cell

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
26
In order of their occurrence what are the steps in RNA interference?
A)Processing of RNA transcript by DGCR8, binding to RISC, transport to cytoplasm, processing by Dicer, binding to target mRNA
B) Processing by Dicer, processing of RNA transcript by DGCR8, transport to cytoplasm, binding to RISC, binding to target mRNA
C)Processing of RNA transcript by DGCR8, transport to cytoplasm, processing by Dicer, binding to RISC, binding to target mRNA
D)Processing of RNA transcript by DGCR8, processing by Dicer, transport to cytoplasm, binding to RISC, binding to target mRNA
A)Processing of RNA transcript by DGCR8, binding to RISC, transport to cytoplasm, processing by Dicer, binding to target mRNA
B) Processing by Dicer, processing of RNA transcript by DGCR8, transport to cytoplasm, binding to RISC, binding to target mRNA
C)Processing of RNA transcript by DGCR8, transport to cytoplasm, processing by Dicer, binding to RISC, binding to target mRNA
D)Processing of RNA transcript by DGCR8, processing by Dicer, transport to cytoplasm, binding to RISC, binding to target mRNA

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
27
The best term for the process by which some exons are included in a mature mRNA from one cell but are absent from the mRNA in another cell is
A) RNA interference.
B) chromosomal remodeling.
C) alternative splicing.
D) Pre-mRNA splicing.
A) RNA interference.
B) chromosomal remodeling.
C) alternative splicing.
D) Pre-mRNA splicing.

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
28
If a human cell has two Barr bodies then the genotype of the individual could be
A) XXXY.
B) XXXXY.
C) XXY.
D) XYY.
A) XXXY.
B) XXXXY.
C) XXY.
D) XYY.

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
29
What is the difference between the trithorax (TrxG) and polycomb (PcG) complexes of proteins?
A)TrxG proteins methylate lysine 27 on histone H3 while PcG complexes methylate lysine 4 on H3
B)Methylation of lysine at position 4 on H3 results in silencing of gene expression
C)TrxG complex proteins activate gene expression by methylating lysine 4 on H3, while PcG complex proteins repress gene expression by methylating lysine 27 on H3.
D)Methylation of H3 on lysine 27 can result in activation of transcription
A)TrxG proteins methylate lysine 27 on histone H3 while PcG complexes methylate lysine 4 on H3
B)Methylation of lysine at position 4 on H3 results in silencing of gene expression
C)TrxG complex proteins activate gene expression by methylating lysine 4 on H3, while PcG complex proteins repress gene expression by methylating lysine 27 on H3.
D)Methylation of H3 on lysine 27 can result in activation of transcription

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
30
In the presence of abundant iron in the cell, the iron binds IRP. The complex then binds to the IREs and
A) an increase in ferritin mRNA translation and a decrease in transferrin receptor mRNA stability.
B) a decrease in transferrin receptor mRNA translation and an increase in ferritin mRNA stability.
C) an increase in ferritin and transferrin receptor mRNA translation.
D) an increase in ferritin and transferrin receptor gene transcription.
A) an increase in ferritin mRNA translation and a decrease in transferrin receptor mRNA stability.
B) a decrease in transferrin receptor mRNA translation and an increase in ferritin mRNA stability.
C) an increase in ferritin and transferrin receptor mRNA translation.
D) an increase in ferritin and transferrin receptor gene transcription.

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
31
Which of the following can control mRNA stability?
A) The stop codon that is used to terminate translation in the mRNA
B) AU rich elements in the mRNA
C) distance of the stop codon to the polyA tail
D) number of A's in polyA tail
A) The stop codon that is used to terminate translation in the mRNA
B) AU rich elements in the mRNA
C) distance of the stop codon to the polyA tail
D) number of A's in polyA tail

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
32
In cancer cells one allele of the tumor suppressor gene p53 is frequently mutated so that the protein is inactive, not produced, or deleted. The other allele will usually have a normal sequence and the promoter remains intact but the gene is not expressed. Sequencing with sodium bisulfite modification of DNA can be used to detect which cytosines are methylated. If the cancer cell DNA is sequenced what would be the anticipated results?
A)Cytosines in or near the promoter region will be methylated
B)Cytosines in or near the promoter will not be methylated
C)Cytosines in the coding region will have an increased mehtylation
D)There will be no differences in the methylation pattern of the promoter of p53 from a cancer cell and a normal cell
A)Cytosines in or near the promoter region will be methylated
B)Cytosines in or near the promoter will not be methylated
C)Cytosines in the coding region will have an increased mehtylation
D)There will be no differences in the methylation pattern of the promoter of p53 from a cancer cell and a normal cell

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
33
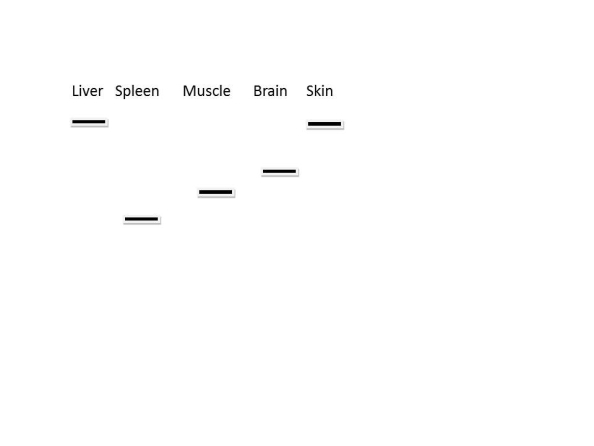 Northern blot analysis can be used to determine the relative amount and size of different RNA transcripts. Above is a representation of a northern blot analysis of several different tissues for the expression of a single gene. Which of the following statements is correct?
Northern blot analysis can be used to determine the relative amount and size of different RNA transcripts. Above is a representation of a northern blot analysis of several different tissues for the expression of a single gene. Which of the following statements is correct?A) Some of the samples contain incompletely processed mRNA
B) Some of the samples contain pre-mRNA
C) The mRNA is differentially spliced
D) All tissues express identical proteins

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
34
It is frequently desirable to experimentally regulate gene regulation and then study the consequences of expressing or suppressing the specific gene. To accomplish this, different gene sequences are fused to promoters or sequences that will be expressed as mRNA. Which of the following would most likely result in regulatable expression?
A) ARE
B) IRE
C) IBP
D) ICR
A) ARE
B) IRE
C) IBP
D) ICR

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
35
Which of the following is part of the process of X chromosome inactivation?
A) Expression of Xist from both chromosomes at the start of the process
B) Binding of multiple Xist transcripts to Xic on the X chromosome that will be inactivated
C) Compaction of the active X chromosme into a Barr body
D) Binding of Tsix transcripts to the X chromosome to be inactivated after the Xist transcripts binds to Xic
A) Expression of Xist from both chromosomes at the start of the process
B) Binding of multiple Xist transcripts to Xic on the X chromosome that will be inactivated
C) Compaction of the active X chromosme into a Barr body
D) Binding of Tsix transcripts to the X chromosome to be inactivated after the Xist transcripts binds to Xic

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck
36
Gene methylation can be detected through the use of restriction endonucleases. Usually these are used in pairs where one enzyme will digest only unmethylated DNA in its recognition sequence while the other is insensitive to methylation. Which of the following statements is correct?
A)The enzyme that is insensitive to methylation serves as a control to make sure the inability of the other enzyme to digest DNA is not due to a mutation.
B)These are used in pairs since experiments have to be replicated and this one way of performing a replication of the experiment
C)The experimental design is flawed since there should be a third enzyme that would serve as a positive cocntrol
D) This experimental design is able to detect most of the methylated C residues in a particular region of DNA.
A)The enzyme that is insensitive to methylation serves as a control to make sure the inability of the other enzyme to digest DNA is not due to a mutation.
B)These are used in pairs since experiments have to be replicated and this one way of performing a replication of the experiment
C)The experimental design is flawed since there should be a third enzyme that would serve as a positive cocntrol
D) This experimental design is able to detect most of the methylated C residues in a particular region of DNA.

Unlock Deck
Unlock for access to all 36 flashcards in this deck.
Unlock Deck
k this deck


