Deck 13: Genomes
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/193
Play
Full screen (f)
Deck 13: Genomes
1
Assembling complete genomic sequences based on the overlap of sequenced fragments can be difficult because the sequenced fragments:
A)may be relatively short (50-100 nucleotides).
B)may have errors in them.
C)may come from either DNA strand.
D)may overlap but come from different locations in the genome.
E)may actually come from bacteria, viruses, or other contaminants.
A)may be relatively short (50-100 nucleotides).
B)may have errors in them.
C)may come from either DNA strand.
D)may overlap but come from different locations in the genome.
E)may actually come from bacteria, viruses, or other contaminants.
A, B, C, D, E
2
Which of the following objects lacks a genome?
A)nucleus
B)chloroplast
C)mitochondrion
D)virus particle
E)All of these choices have a genome.
A)nucleus
B)chloroplast
C)mitochondrion
D)virus particle
E)All of these choices have a genome.
E
3
Consider the sentence fragments shown in Figure 13.1, below. 1 "It has not escaped our no
2 specific pairing we have post
3 suggests a plausible copying
4 d our notice that the specifi
5 ve postulated immediately suggests
6 pying mechanism for the genetic material." Which of the following would make assembling the sentence more difficult?
A)If each of the words in the sentence were used only once.
B)If some of the words in the sentence were used several times.
C)If the fragments were longer.
D)If the letters were randomly assembled and did not form known words.
2 specific pairing we have post
3 suggests a plausible copying
4 d our notice that the specifi
5 ve postulated immediately suggests
6 pying mechanism for the genetic material." Which of the following would make assembling the sentence more difficult?
A)If each of the words in the sentence were used only once.
B)If some of the words in the sentence were used several times.
C)If the fragments were longer.
D)If the letters were randomly assembled and did not form known words.
If some of the words in the sentence were used several times.
If the letters were randomly assembled and did not form known words.
If the letters were randomly assembled and did not form known words.
4
Sometimes a single-stranded molecule of RNA is able to fold back on itself because the nucleotide sequence on one part of the RNA is complementary to another part. This sequence motif results in a:
A)transcription factor binding site.
B)hairpin-shaped structure.
C)chromosome scaffold.
D)negative supercoil.
E)positive supercoil.
A)transcription factor binding site.
B)hairpin-shaped structure.
C)chromosome scaffold.
D)negative supercoil.
E)positive supercoil.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
5
Using current DNA-sequencing technology, it is possible to sequence an entire chromosome (e.g., human chromosome 1, which contains approximately 250 million nucleotides) as one long molecule.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
6
A shotgun sequencing project yields the following sequence for a strand of DNA. 5-GGTTTGGAGTAT-3
In assembling the genome sequence from the short fragments, you look for other fragments that overlap the sequence above. Which of the following sequences overlaps with the sequence above?
A)5-GGTTTGGAGTGG-3
B)5-AATTTGGAGTAT-3
C)5-TTTGGAGTATGG-3
D)All of the sequences overlap with sequence above.
E)None of the sequences overlaps with the sequence above.
In assembling the genome sequence from the short fragments, you look for other fragments that overlap the sequence above. Which of the following sequences overlaps with the sequence above?
A)5-GGTTTGGAGTGG-3
B)5-AATTTGGAGTAT-3
C)5-TTTGGAGTATGG-3
D)All of the sequences overlap with sequence above.
E)None of the sequences overlaps with the sequence above.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
7
A shotgun sequencing project yields the following sequence for a strand of DNA. 5'-GGTTTGGAGTAT-3'
Which of the following sequences overlaps with the sequence above?
A)5-GGTTTTTAGACT-3
B)5-GTGTTGGAGCTT-3
C)5-GAGTATCCAAAT-3
D)5-GGCTTGAGGTTA-3
E)None of the sequences overlaps with the sequence above.
Which of the following sequences overlaps with the sequence above?
A)5-GGTTTTTAGACT-3
B)5-GTGTTGGAGCTT-3
C)5-GAGTATCCAAAT-3
D)5-GGCTTGAGGTTA-3
E)None of the sequences overlaps with the sequence above.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
8
A shotgun genome sequencing project is designed in which the average length of a sequenced fragment is 100 nucleotides and each nucleotide is to be sequenced an average of 30 times. For the human genome, which is 3 × 109 base pairs long, how many fragments of 100 nucleotides would need to be sequenced?
A)(3 × 109)× 30 × 100 = 9 × 1011
B)(3 × 109)× 30 ÷ 100 = 9 × 108
C)(3 × 109)× 100 ÷ 30 = 1 × 1010
D)(3 × 109)÷ (30 × 100)= 1 × 106
A)(3 × 109)× 30 × 100 = 9 × 1011
B)(3 × 109)× 30 ÷ 100 = 9 × 108
C)(3 × 109)× 100 ÷ 30 = 1 × 1010
D)(3 × 109)÷ (30 × 100)= 1 × 106

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
9
A shotgun genome sequencing project is designed in which the average length of a sequenced fragment is 150 nucleotides and each nucleotide is to be sequenced an average of 20 times. For a genome that is 3 × 107 base pairs long, how many fragments of 150 nucleotides would need to be sequenced?
A)(3 × 107)× 20 × 150 = 9 × 1010
B)(3 × 107)× 150 ÷ 20 = 2.25 × 108
C)(3 × 107)÷ (20 × 150)= 1 × 104
D)(3 × 107)× 20 ÷ 150 = 4 × 106
A)(3 × 107)× 20 × 150 = 9 × 1010
B)(3 × 107)× 150 ÷ 20 = 2.25 × 108
C)(3 × 107)÷ (20 × 150)= 1 × 104
D)(3 × 107)× 20 ÷ 150 = 4 × 106

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
10
Sequences that are conserved (i.e., similar in many different organisms) are unlikely to be functionally important.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
11
Repeated DNA sequences represent a special challenge in genome sequence assembly. Which of the following would be harder to assemble correctly, assuming the number of copies of the repeat can be determined?
A)sequences containing repeats longer than the DNA fragments to assemble
B)sequences containing repeats shorter than the DNA fragments to assemble
A)sequences containing repeats longer than the DNA fragments to assemble
B)sequences containing repeats shorter than the DNA fragments to assemble

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
12
Figure 13.3 below provides examples of various types of sequences that can be found in a segment of double-stranded DNA. Which one of the following is MOST likely to be protein-coding? 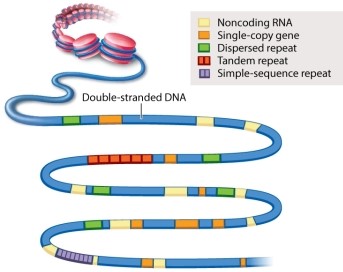
A)noncoding RNA
B)single-copy gene
C)dispersed repeat
D)tandem repeat
E)simple-sequence repeat

A)noncoding RNA
B)single-copy gene
C)dispersed repeat
D)tandem repeat
E)simple-sequence repeat

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
13
Why is a DNA sequence motif consisting of codons uninterrupted by a stop codon often called a "putative" open reading frame instead of just an open reading frame?
A)because it could be due to chance
B)because it could be due to sequencing error
C)because it might not be transcribed
D)because it could be part of an intron
E)All of these choices are correct.
A)because it could be due to chance
B)because it could be due to sequencing error
C)because it might not be transcribed
D)because it could be part of an intron
E)All of these choices are correct.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
14
Which one of the following steps comes FIRST in shotgun sequencing?
A)breaking the DNA into small fragments
B)putting the sequences in the correct order
C)matching regions of overlap
D)sequencing the DNA
E)reconstructing the long sequence of nucleotides
A)breaking the DNA into small fragments
B)putting the sequences in the correct order
C)matching regions of overlap
D)sequencing the DNA
E)reconstructing the long sequence of nucleotides

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
15
In using the shotgun approach to genome sequencing, the sequenced fragments originate from:
A)the beginning of a gene.
B)the center of a gene.
C)random sites scattered across the genome.
D)targeted sites scattered across the genome.
E)None of the other answer options is correct.
A)the beginning of a gene.
B)the center of a gene.
C)random sites scattered across the genome.
D)targeted sites scattered across the genome.
E)None of the other answer options is correct.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
16
If a genome is sequenced in such a way that any given nucleotide is present in n sequenced fragments, then if each nucleotide is equally likely to be sequenced, the expected proportion of nucleotides that are not present in any of the fragments is given by e-n, where e is the base of natural logarithms. (The value of e is approximately 2.7182.) What is the probability that a nucleotide is not sequenced if n = 1? If n = 2? If n = 3?

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
17
Approximately what percentage of the human genome actually codes for proteins?
A)1)0%
B)2)5%
C)45.0%
D)97.5%
E)99.0%
A)1)0%
B)2)5%
C)45.0%
D)97.5%
E)99.0%

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
18
Genome sequencing includes the following steps:
1) putting the sequences in the correct order
2) matching regions of overlap
3) breaking the DNA into small fragments
4) reconstructing the long sequence of nucleotides
"5) sequencing the DNA
In what order are these steps carried out?"
A)1-2-3-4-5
B)5-2-3-1-4
C)3-5-2-1-4
D)3-5-2-4-1
E)5-4-3-2-1
1) putting the sequences in the correct order
2) matching regions of overlap
3) breaking the DNA into small fragments
4) reconstructing the long sequence of nucleotides
"5) sequencing the DNA
In what order are these steps carried out?"
A)1-2-3-4-5
B)5-2-3-1-4
C)3-5-2-1-4
D)3-5-2-4-1
E)5-4-3-2-1

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
19
What could be one way to solve the problem of tandem repeats complicating the alignment of DNA fragments in shotgun sequencing?
A)develop techniques to sequence longer DNA fragments
B)sequence more DNA fragments
C)combine sequencing with chromosome painting
D)sequence both strands of the tandem repeats
A)develop techniques to sequence longer DNA fragments
B)sequence more DNA fragments
C)combine sequencing with chromosome painting
D)sequence both strands of the tandem repeats

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
20
A repetitive sequence that is 300 base pairs in length is present in 200 identical copies arranged end to end in one region of the genome. Could you assemble short sequences across this region? Explain.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
21
Knowing an individual's DNA sequence may be beneficial because it makes it possible to predict:
A)susceptibility to disease.
B)response to medications.
C)physical differences.
D)the occurrence of a disease.
E)All of these choices are correct.
A)susceptibility to disease.
B)response to medications.
C)physical differences.
D)the occurrence of a disease.
E)All of these choices are correct.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
22
In the eukaryotic DNA sequence below, each highlighted sequence consists of a simple-sequence repeat. How are the underlined regions organized relative to one another? ATTATCATCATCATCATCATTTACTAATCCTCATCATCATCATCATGGAATCTACATCATCATCATCAT
A)as tandem repeats
B)as dispersed repeats
C)as inverted repeats
D)as long terminal repeats
E)as short terminal repeats
A)as tandem repeats
B)as dispersed repeats
C)as inverted repeats
D)as long terminal repeats
E)as short terminal repeats

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
23
A baby was born whose biological mother and father each donated a set of chromosomes, and an unrelated woman donated the mitochondria. How many "genomes" did this child inherit? How many "genomes" does the child have?
A)1; 1
B)2; 1
C)2)5; 2.5
D)3; 1
E)3; 3
A)1; 1
B)2; 1
C)2)5; 2.5
D)3; 1
E)3; 3

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
24
Which one of the following pairs of people has the exact same genome?
A)father and son
B)mother and daughter
C)identical twins
D)fraternal twins
E)None of the other answer options is correct.
A)father and son
B)mother and daughter
C)identical twins
D)fraternal twins
E)None of the other answer options is correct.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
25
Which one of the following statements about an open reading frame is TRUE?
A)An open reading frame consists of a long string of codons for amino acids.
B)A long open reading frame may be protein coding.
C)An open reading frame is uninterrupted by a stop codon.
D)All of these choices are correct.
A)An open reading frame consists of a long string of codons for amino acids.
B)A long open reading frame may be protein coding.
C)An open reading frame is uninterrupted by a stop codon.
D)All of these choices are correct.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
26
"Shotgun" sequencing involves aligning many small sequences from a genome based on sequence similarities.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
27
A possible negative consequence of personalized medicine could be:
A)predicting responses to medications.
B)incorrect predictions of sequence motifs.
C)difficulty obtaining insurance if susceptibility to disease becomes public information.
D)tailoring treatments to an individual.
E)None of the other answer options represents negative consequences of personalized medicine.
A)predicting responses to medications.
B)incorrect predictions of sequence motifs.
C)difficulty obtaining insurance if susceptibility to disease becomes public information.
D)tailoring treatments to an individual.
E)None of the other answer options represents negative consequences of personalized medicine.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
28
Sequence assembly is accomplished by aligning the fragments:
A)in the laboratory by running them through gel electrophoresis.
B)on paper.
C)by using a complex computer program.
D)by joining histones.
E)None of the other answer options is correct.
A)in the laboratory by running them through gel electrophoresis.
B)on paper.
C)by using a complex computer program.
D)by joining histones.
E)None of the other answer options is correct.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
29
Which one of the following types of sequences can be found in a genome?
A)regulatory elements of protein-coding genes
B)noncoding introns
C)protein-coding exons
D)coding sequence for RNAs
E)All of these choices are correct.
A)regulatory elements of protein-coding genes
B)noncoding introns
C)protein-coding exons
D)coding sequence for RNAs
E)All of these choices are correct.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
30
The human chromosome 1 is approximately 250 million nucleotides long. In order to sequence human chromosome 1, the chromosome is broken up into hundreds of small fragments. These short fragments are placed together in the correct order to generate the long, continuous sequence of nucleotides in chromosome 1. Placing these fragments together is referred to as sequence:
A)sequencing.
B)shuffling.
C)assembly.
D)numbering.
E)assignment.
A)sequencing.
B)shuffling.
C)assembly.
D)numbering.
E)assignment.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
31
Knowledge of your personal genome:
A)tells you what genetic diseases you will get and when in life they will appear.
B)identifies genetic risk factors for disease.
C)allows you to make informed decisions about health.
D)tells you which risky lifestyle choices are OK for you.
A)tells you what genetic diseases you will get and when in life they will appear.
B)identifies genetic risk factors for disease.
C)allows you to make informed decisions about health.
D)tells you which risky lifestyle choices are OK for you.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
32
Repeated sequences within eukaryotic genomes may be represented as long repeated sequences next to each other, and are called _____ repeats. Repeated sequences in eukaryotic genomes can also be spread throughout the genome, and are called _____ repeats.
A)tandem; dispersed
B)dispersed; tandem
C)overlapping; adjacent
D)adjacent; overlapping
E)None of the other answer options is correct.
A)tandem; dispersed
B)dispersed; tandem
C)overlapping; adjacent
D)adjacent; overlapping
E)None of the other answer options is correct.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
33
All of the genetic material transferred from a parent to an offspring is known as a:
A)karyotype.
B)genome.
C)open reading frame.
D)nucleoid.
E)mRNA.
A)karyotype.
B)genome.
C)open reading frame.
D)nucleoid.
E)mRNA.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
34
What was the goal of the Human Genome Project?
A)identifying every gene in the human sex cell
B)identifying every gene in the human somatic cell
C)sequencing every protein in the human sex cell
D)identifying every protein in the human somatic cell
E)sequencing every gene in the human cell
A)identifying every gene in the human sex cell
B)identifying every gene in the human somatic cell
C)sequencing every protein in the human sex cell
D)identifying every protein in the human somatic cell
E)sequencing every gene in the human cell

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
35
Which of the following statements is TRUE regarding a genome?
A)Genomes always contain DNA.
B)Genomes are composed of chromosomes.
C)Genomes are "heritable," or passed from parents to offspring.
D)Only prokaryotic and eukaryotic organisms possess genomes.
E)All of these choices are correct.
A)Genomes always contain DNA.
B)Genomes are composed of chromosomes.
C)Genomes are "heritable," or passed from parents to offspring.
D)Only prokaryotic and eukaryotic organisms possess genomes.
E)All of these choices are correct.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
36
Except for twins and other multiple births, the genome sequence is different in every individual.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
37
Repeated sequences make genome sequencing challenging for of all of the following reasons EXCEPT:
A)repeated sequences are typically longer than the short sequences obtained by automated sequencing.
B)there is no easy way to determine how many copies of the same repeat are present within one chromosome.
C)short repeats may fold back upon themselves to form a double-stranded structure that is not easily sequenced.
D)the long repeats often have mutations that are not easily sequenced.
E)All of these choices are correct.
A)repeated sequences are typically longer than the short sequences obtained by automated sequencing.
B)there is no easy way to determine how many copies of the same repeat are present within one chromosome.
C)short repeats may fold back upon themselves to form a double-stranded structure that is not easily sequenced.
D)the long repeats often have mutations that are not easily sequenced.
E)All of these choices are correct.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
38
Which one of the following represents a consequence of sequencing an organism's genome?
A)An individual's "personal genome" can be sequenced and compared to the species standard to determine potential for disease inheritance.
B)Genome sequences can be compared among species to determine evolutionary relationships.
C)A sequenced genome can be annotated and searched for open reading frames.
D)Once an organism's genome is sequenced, everything is known about that organism's protein expression and behavior.
A)An individual's "personal genome" can be sequenced and compared to the species standard to determine potential for disease inheritance.
B)Genome sequences can be compared among species to determine evolutionary relationships.
C)A sequenced genome can be annotated and searched for open reading frames.
D)Once an organism's genome is sequenced, everything is known about that organism's protein expression and behavior.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
39
Analysis of similarities and differences in the genomes of different species is referred to as:
A)sequence assembly.
B)genome annotation.
C)comparative genomics.
D)personalized medicine.
E)genome conservation.
A)sequence assembly.
B)genome annotation.
C)comparative genomics.
D)personalized medicine.
E)genome conservation.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
40
An individual's genome sequence can reveal his or her disease susceptibilities and drug sensitivities, which allows treatments to be tailored to the individual. This approach is called:
A)personalized medicine.
B)sequence assembly.
C)shotgun sequencing.
D)genome annotation.
E)None of the answer options is correct.
A)personalized medicine.
B)sequence assembly.
C)shotgun sequencing.
D)genome annotation.
E)None of the answer options is correct.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
41
Which of the following statements are TRUE of the reference human genome sequence that was announced in the year 2000?
A)It represents what the genome of a perfectly healthy person would be.
B)It does not really exist in any human being.
C)It corresponds to the genome sequence of the common ancestor of all humans.
D)includes no mutant genes.
E)It is accessible online as public information.
A)It represents what the genome of a perfectly healthy person would be.
B)It does not really exist in any human being.
C)It corresponds to the genome sequence of the common ancestor of all humans.
D)includes no mutant genes.
E)It is accessible online as public information.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
42
A certain sequence motif consists of 5 nucleotide pairs. In a random sequence of double-stranded DNA in which each nucleotide is equally likely to have any of the four bases, what is the probability that any group of 5 adjacent nucleotides has the same sequence as the sequence motif?
A)(1/5)2 = 1/25
B)(1/5)4 = 1/625
C)(1/2)5 = 1/32
D)(1/4)5 = 1/1024
E)None of the answer options is correct.
A)(1/5)2 = 1/25
B)(1/5)4 = 1/625
C)(1/2)5 = 1/32
D)(1/4)5 = 1/1024
E)None of the answer options is correct.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
43
In a genome in which the mean distance between occurrences of a certain sequence motif is 1024 base pairs, about half of the distances between occurrences are smaller than 700 base pairs. How is it possible that more than half of the observations are smaller than the mean?

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
44
In a random sequence of double-stranded DNA, the probability that there are exactly n nucleotide pairs between two successive occurrences of a short sequence motif is given by (1 - x)nx, where x is the probability of occurrence of the motif along the sequence. When x = 1/512, the mean distance between occurrences is 512 nucleotide pairs. What is the probability that the distance between two occurrences is exactly equal to this mean?

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
45
In a random sequence of double-stranded DNA, the probability that there are n or fewer nucleotide pairs between two successive occurrences of a short sequence motif is given by 1 - (1 - x)n, where x is the probability of occurrence of the motif along the sequence. When x = 1/512, the mean distance between occurrences is 512 nucleotide pairs. What is the probability that the distance between two occurrences is smaller than the mean?

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
46
In the problem above, what is the value of n for which half the distances between occurrences are < n and half are > n?

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
47
Which one of the following is a feature of DNA that could allow you to distinguish between a DNA sequence that is protein coding and a DNA sequence that is not?
A)Protein-coding DNA sequences contain U (Uracil), and nontranscribed sequences contain T (Thymine).
B)Protein-coding regions frequently contain long open reading frames; others rarely do.
C)Protein-coding sequences are sets of amino acids; others are sets of nucleotides.
D)Protein-coding sequences are single stranded; others sequences are double stranded.
E)All of these choices are correct.
A)Protein-coding DNA sequences contain U (Uracil), and nontranscribed sequences contain T (Thymine).
B)Protein-coding regions frequently contain long open reading frames; others rarely do.
C)Protein-coding sequences are sets of amino acids; others are sets of nucleotides.
D)Protein-coding sequences are single stranded; others sequences are double stranded.
E)All of these choices are correct.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
48
Which of the following would be found in DNA sequences but not in mRNA sequences?
A)transcription start sites
B)translation start sites
C)open reading frames
D)exons
E)All of these choices are correct.
A)transcription start sites
B)translation start sites
C)open reading frames
D)exons
E)All of these choices are correct.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
49
The eukaryotic DNA sequence below is a tandem repeat. It is also what other kind of repeat? CACACACACACACACACACACACACACACACACACACA
A)transposon repeat
B)dispersed repeat
C)simple sequence repeat
D)long terminal repeat
E)short terminal repeat
A)transposon repeat
B)dispersed repeat
C)simple sequence repeat
D)long terminal repeat
E)short terminal repeat

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
50
In a random sequence of double-stranded DNA with equal nucleotide frequencies, what is the average number of nucleotide pairs between two occurrences of a sequence motif consisting of four nucleotides?
A)16
B)32
C)64
D)128
E)256
A)16
B)32
C)64
D)128
E)256

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
51
Which one of the following statements about transposable elements is CORRECT?
A)They replicate themselves and insert themselves into new positions.
B)They are sometimes described as selfish and called the ultimate parasite.
C)They make up 45% of the DNA in the human genome.
D)They can transpose via DNA replication or an RNA intermediate.
E)All of these choices are correct.
A)They replicate themselves and insert themselves into new positions.
B)They are sometimes described as selfish and called the ultimate parasite.
C)They make up 45% of the DNA in the human genome.
D)They can transpose via DNA replication or an RNA intermediate.
E)All of these choices are correct.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
52
Whole genome sequencing is often approached by a shotgun sequencing technique in which large genomes are:
A)randomly digested, short fragments are sequenced, and the overlapping sequences are assembled in order.
B)digested in a few specific spots, large fragments are sequenced, and the overlapping sequences are assembled in order.
C)randomly digested, short fragments are sequenced, and the overlapping sequences are assembled from largest to smallest.
D)randomly digested, short fragments are sequenced, and the overlapping sequences are assembled from smallest to largest.
E)All of these choices are correct.
A)randomly digested, short fragments are sequenced, and the overlapping sequences are assembled in order.
B)digested in a few specific spots, large fragments are sequenced, and the overlapping sequences are assembled in order.
C)randomly digested, short fragments are sequenced, and the overlapping sequences are assembled from largest to smallest.
D)randomly digested, short fragments are sequenced, and the overlapping sequences are assembled from smallest to largest.
E)All of these choices are correct.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
53
According to the phylogenetic tree showing evolutionary relationships among viruses in Figure 13.6 below, which one of the following is MOST closely related to the human lentivirus HIV2? 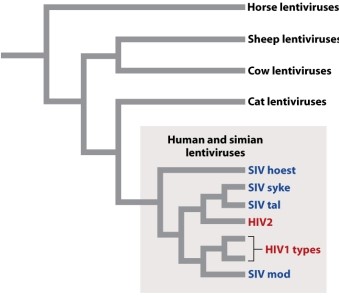
A)a horse lentivirus
B)human lentivirus HIV1
C)a cow lentivirus
D)a simian lentivirus
E)a cat lentivirus

A)a horse lentivirus
B)human lentivirus HIV1
C)a cow lentivirus
D)a simian lentivirus
E)a cat lentivirus

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
54
Consider a stretch of DNA with two short sequence motifs separated by an unknown number of base pairs. Each position in the intervening DNA is equally likely to be any of the four nucleotides. Would you expect many sites to be close together and a few farther apart? Few to be close together and many farther apart? Equal spacing? A normal distribution of distances between sites? In other words, which of the following curves do you think BEST approximates the distribution of the number of nucleotide pairs between two occurrences of a short sequence motif? 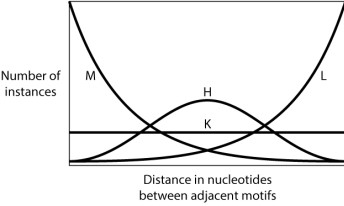
A)Curve M
B)Curve H
C)Curve K
D)Curve L

A)Curve M
B)Curve H
C)Curve K
D)Curve L

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
55
If you want to identify protein-coding sequences, why is it more efficient to study messenger RNAs than DNA?
A)Because mRNAs have poly-A tails
B)Because mRNAs do not contain the promoter sequence
C)Because triplets of nucleotides in DNA do not indicate what amino acids will be present in a protein.
D)Because processed mRNA does not contain introns.
A)Because mRNAs have poly-A tails
B)Because mRNAs do not contain the promoter sequence
C)Because triplets of nucleotides in DNA do not indicate what amino acids will be present in a protein.
D)Because processed mRNA does not contain introns.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
56
Which of the following represent repeated sequences commonly found in the human genome?
A)long sequences repeated in tandem
B)long sequences dispersed throughout the genome
C)short sequences repeated many times in tandem
D)sequences whose transcript can fold back to form a hairpin
E)All of these choices are correct.
A)long sequences repeated in tandem
B)long sequences dispersed throughout the genome
C)short sequences repeated many times in tandem
D)sequences whose transcript can fold back to form a hairpin
E)All of these choices are correct.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
57
In the eukaryotic DNA sequence below, which type of repeat is underlined? ATTATTTACTAATCCTCATCATCATCATCATGGAATTCATAATGCTAATGG
A)tandem repeat
B)dispersed repeat
C)simple sequence repeat
D)long terminal repeat
E)short terminal repeat
A)tandem repeat
B)dispersed repeat
C)simple sequence repeat
D)long terminal repeat
E)short terminal repeat

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
58
Sequences of genomic DNA, and its corresponding messenger RNA (mRNA), are often compared to obtain valuable information for genome annotation. Why is this comparison useful?
A)The open reading frame of the mRNA includes the introns of the genomic DNA.
B)The exclusion of introns in mRNA reveals the intron-exon structure of many protein coding genes.
C)The genomic DNA is longer because the exons are spliced together.
D)The genomic DNA is shorter because the exons are spliced together.
E)The sequences of genomic DNA and mRNA are identical, which serves as independent validation.
A)The open reading frame of the mRNA includes the introns of the genomic DNA.
B)The exclusion of introns in mRNA reveals the intron-exon structure of many protein coding genes.
C)The genomic DNA is longer because the exons are spliced together.
D)The genomic DNA is shorter because the exons are spliced together.
E)The sequences of genomic DNA and mRNA are identical, which serves as independent validation.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
59
In addition to noncoding sequences, most eukaryotic genomes contain not only functional genes that encode proteins, but also nonfunctional pseudogenes. Pseudogenes are copies of functional genes, but contain mutations that prevent function or could block either transcription or translation. Which of the following could indicate that a gene-like DNA sequence is a pseudogene sequence and not a functioning gene?
A)The sequence has no transcription-factor binding sites.
B)The sequence has no putative open reading frame.
C)The sequence is not transcribed.
D)The sequence has introns and spliceosome-binding sites.
E)The sequence folds back on itself to form a hairpin.
A)The sequence has no transcription-factor binding sites.
B)The sequence has no putative open reading frame.
C)The sequence is not transcribed.
D)The sequence has introns and spliceosome-binding sites.
E)The sequence folds back on itself to form a hairpin.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
60
Which one of the following BEST describes why genome sequencing can be complicated by repeated sequences?
A)The repeated sequences are often longer than the sequences obtained by automated sequencing.
B)Automated sequencing devices interpret repeated sequences as a single-copy sequence.
C)Repeated sequences artificially inflate the genome size, leading to an exaggerated interpretation of the complexity of the organism.
D)The repeated sequences are too small to gather usable sequence information from.
E)All of these choices are correct.
A)The repeated sequences are often longer than the sequences obtained by automated sequencing.
B)Automated sequencing devices interpret repeated sequences as a single-copy sequence.
C)Repeated sequences artificially inflate the genome size, leading to an exaggerated interpretation of the complexity of the organism.
D)The repeated sequences are too small to gather usable sequence information from.
E)All of these choices are correct.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
61
A researcher annotating the rabbit genome would describe which of the following?
A)noncoding DNA
B)tandem or dispersed repeats
C)intron/exon boundaries in genes
D)DNA encoding hairpin RNAs
E)All of these choices are correct.
A)noncoding DNA
B)tandem or dispersed repeats
C)intron/exon boundaries in genes
D)DNA encoding hairpin RNAs
E)All of these choices are correct.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
62
Which one of following statements explains why genome annotation is an ongoing, dynamic process in need of continued updating?
A)Our understanding of genes and gene function changes.
B)The functions and interactions of macromolecules are never the same.
C)Sequences change daily because mutation rates are extremely high.
D)Certain structures like hairpins are not predictable using older annotation techniques.
E)All of these choices are correct.
A)Our understanding of genes and gene function changes.
B)The functions and interactions of macromolecules are never the same.
C)Sequences change daily because mutation rates are extremely high.
D)Certain structures like hairpins are not predictable using older annotation techniques.
E)All of these choices are correct.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
63
Which of the following is NOT a type of sequence identified by gene annotation?
A)topoisomerase
B)noncoding RNA
C)single copy gene
D)tandem repeat
E)simple sequence repeat
A)topoisomerase
B)noncoding RNA
C)single copy gene
D)tandem repeat
E)simple sequence repeat

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
64
How can researchers distinguish exons from introns in a segment of DNA?
A)Only exons contain three-base sequences that can code for amino acids.
B)Primers won't bind to introns.
C)Exons have a characteristic sequence.
D)The sequence of exons complements mRNA molecules in the cell.
A)Only exons contain three-base sequences that can code for amino acids.
B)Primers won't bind to introns.
C)Exons have a characteristic sequence.
D)The sequence of exons complements mRNA molecules in the cell.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
65
A small infectious agent that contains a nucleic acid genome packaged inside a protein coat is called a(n):
A)bacterium.
B)virus.
C)tRNA.
D)archaeon.
A)bacterium.
B)virus.
C)tRNA.
D)archaeon.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
66
The genome of a virus can contain which of the following?
A)genes that code for surface proteins
B)genes that code for components of the capsid
C)genes that code for reverse transcriptase
D)genes that code for proteases
E)All of these choices are correct.
A)genes that code for surface proteins
B)genes that code for components of the capsid
C)genes that code for reverse transcriptase
D)genes that code for proteases
E)All of these choices are correct.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
67
In HIV, RNA is transcribed to DNA by:
A)the matrix.
B)reverse transcriptase.
C)surface glycoprotein.
D)the capsid.
E)integrase.
A)the matrix.
B)reverse transcriptase.
C)surface glycoprotein.
D)the capsid.
E)integrase.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
68
Imagine that a researcher is comparing the sequence of several protein-coding genes among mice, rabbits, and humans. She finds that, for most of these genes, the rabbit sequences are more similar to the human sequences than are the mouse sequences. What can she deduce?
A)Mice and rabbits do not share a common ancestor.
B)Mice and humans share a more "recent" common ancestor than do rabbits and humans.
C)No similar proteins exist in mice and humans.
D)More sequences are conserved between rabbits and humans than between humans and mice.
E)Humans and mice do not share a common ancestor.
A)Mice and rabbits do not share a common ancestor.
B)Mice and humans share a more "recent" common ancestor than do rabbits and humans.
C)No similar proteins exist in mice and humans.
D)More sequences are conserved between rabbits and humans than between humans and mice.
E)Humans and mice do not share a common ancestor.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
69
The MOST likely origin of human HIV1 virus is a related virus that infects:
A)horses.
B)sheep.
C)cows.
D)cats.
E)chimpanzees.
A)horses.
B)sheep.
C)cows.
D)cats.
E)chimpanzees.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
70
The coding region of mRNA is called an _____, whereas the noncoding region is called a(n) _____.
A)intron; exon
B)exon; intron
C)exon; open reading frame
D)intron; sequence motif
E)intron; transposable element
A)intron; exon
B)exon; intron
C)exon; open reading frame
D)intron; sequence motif
E)intron; transposable element

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
71
Imagine a genomic researcher who is analyzing the genome of different types of cats. She finds that a particular sequence in the North American Bobcat genome is exactly identical to a sequence found in the common house cat, while most other sequences in those two genomes differ at many nucleotides. (The most recent common ancestor between bobcats and house cats is estimated to be about 6.8 million years, plenty of time for mutation to generate DNA sequence variation.) Which of the following could explain the identical sequence in these otherwise differing genomes?
A)The sequence encodes a gene that is critical for life and cannot be easily mutated while retaining function.
B)The sequence is contained in a retrovirus that has infected both species.
C)The sequence encodes a protein critical for the production of fur color.
D)The sequence is from an intron of a gene that encodes a muscle protein.
A)The sequence encodes a gene that is critical for life and cannot be easily mutated while retaining function.
B)The sequence is contained in a retrovirus that has infected both species.
C)The sequence encodes a protein critical for the production of fur color.
D)The sequence is from an intron of a gene that encodes a muscle protein.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
72
When an open reading frame (ORF) is identified, it may not actually correspond to the amino acid sequence of any polypeptide in the cell. Why not?
A)The DNA may not be transcribed into RNA.
B)It may contain one or more stop codons in the middle.
C)The ORF could be due to chance.
D)The amino acid sequence may not match any known sequence.
A)The DNA may not be transcribed into RNA.
B)It may contain one or more stop codons in the middle.
C)The ORF could be due to chance.
D)The amino acid sequence may not match any known sequence.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
73
When a genome is annotated, researchers identify all of the protein-coding genes and assign each protein with a function.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
74
Genomic pattern recognition begins with identification of sequence _____, which are telltale signs that identify the type of sequence it is.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
75
Which of the following statements is TRUE regarding sequence motifs?
A)Sequence motifs are found in DNA but not in RNA.
B)Sequence motifs in DNA are found only upstream of protein-coding regions.
C)An open reading frame is a type of sequence motif.
D)Sequence motifs in RNA provide no information about sequence motifs in DNA.
E)None of the answer options is correct.
A)Sequence motifs are found in DNA but not in RNA.
B)Sequence motifs in DNA are found only upstream of protein-coding regions.
C)An open reading frame is a type of sequence motif.
D)Sequence motifs in RNA provide no information about sequence motifs in DNA.
E)None of the answer options is correct.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
76
The reverse transcriptase in the HIV genome encodes a protein that:
A)makes RNA from protein.
B)makes RNA from DNA.
C)makes protein from DNA.
D)makes tRNA from RNA.
E)makes DNA from RNA.
A)makes RNA from protein.
B)makes RNA from DNA.
C)makes protein from DNA.
D)makes tRNA from RNA.
E)makes DNA from RNA.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
77
All DNA sequences are transcribed into RNA.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
78
_____ is the process by which researchers identify various types of sequences.
A)Genome annotation
B)The C-value paradox
C)Karyotype synthesis
D)Chromosome condensation
E)Comparative genomics
A)Genome annotation
B)The C-value paradox
C)Karyotype synthesis
D)Chromosome condensation
E)Comparative genomics

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
79
Comparative _____ is the analysis of the similarities and differences in protein-coding genes and other types of sequences in the genomes of different species.
A)genomics
B)evolution
C)speciation
D)annotation
E)proteomics
A)genomics
B)evolution
C)speciation
D)annotation
E)proteomics

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck
80
Why does Figure 13.6 (below) suggest that human HIV came from chimpanzees at least twice? 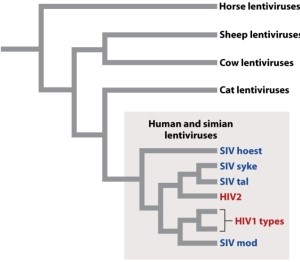
A)Because not all human HIVs are clustered together in the tree.
B)Because they are both more evolved than cat viruses.
C)Because they all share one common ancestor.
D)Because horse viruses are more distantly related.

A)Because not all human HIVs are clustered together in the tree.
B)Because they are both more evolved than cat viruses.
C)Because they all share one common ancestor.
D)Because horse viruses are more distantly related.

Unlock Deck
Unlock for access to all 193 flashcards in this deck.
Unlock Deck
k this deck


