
Genetics: Analysis and Principles 5th Edition by Robert Brooker
Edition 5ISBN: 978-0073525341
Genetics: Analysis and Principles 5th Edition by Robert Brooker
Edition 5ISBN: 978-0073525341 Exercise 23
Membrane proteins often have transmembrane regions that span the membrane in an -helical conformation. These transmembrane segments are about 20 amino acids long and usually contain amino acids with nonpolar (i.e., hydrophobic) amino acid side chains. Researchers can predict whether a polypeptide sequence has transmembrane segments based on the occurrence of segments that contain 20 nonpolar amino acids. To do so, each amino acid is assigned a hydropathy value, based on the chemistry of its amino acid side chain. Amino acids with very nonpolar side chains are given a high (positive) value, whereas amino acids that are charged and/or polar are given low (negative) values. The hydropathy values usually range from about +4 to -4.Computer programs have been devised that scan the amino acid sequence of a polypeptide and calculate values based on the hydropathy values of the amino acid side chains. The program usually scans a window of seven amino acids and assigns an average hydropathy value. For example, the program would scan amino acids 1-7 and give an average value, then it would scan 2-8 and give a value, then it would scan 3-9 and give a value, and so on, until it reached the end of the polypeptide sequence (i.e., until it reached the carboxyl-terminus).The program then produces a figure, known as a hydropathy plot, which describes the average hydropathy values throughout the entire polypeptide sequence. An example of a hydropathy plot is shown here. 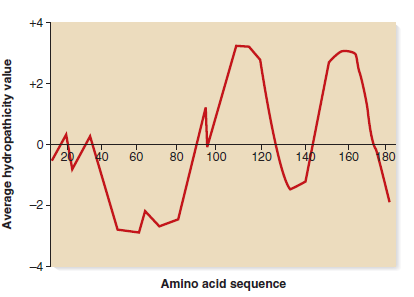
a. How many transmembrane segments are likely in this polypeptide
B. Draw the structure of this polypeptide if it were embedded in the plasma membrane. Assume that the amino terminus is found in the cytoplasm of the cell.

a. How many transmembrane segments are likely in this polypeptide
B. Draw the structure of this polypeptide if it were embedded in the plasma membrane. Assume that the amino terminus is found in the cytoplasm of the cell.
Explanation
Genetics: Analysis and Principles 5th Edition by Robert Brooker
Why don’t you like this exercise?
Other Minimum 8 character and maximum 255 character
Character 255


