Deck 13: Chromosomal Rearrangements and Changes in Chromosome Number
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/49
Play
Full screen (f)
Deck 13: Chromosomal Rearrangements and Changes in Chromosome Number
1
Choose the phrase from the right column that best fits the term in the left column. 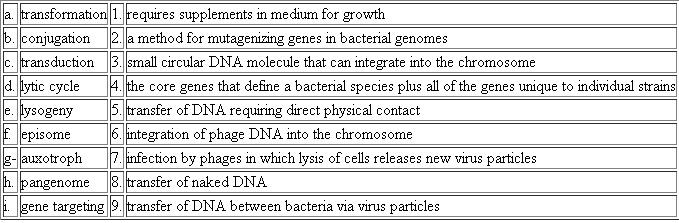

(a)Transformation is a process in which bacterium takes up DNA (deoxyribonucleic acid) fragments spontaneously from their surroundings. These fragments are naked DNA because there is no contact between two bacteria and these fragments are freely present in surrounding of the bacterium. Hence, we can say there is the transfer of naked DNA from donor to a recipient cell.(b)Conjugation is a gene transfer process in which DNA fragments transferred from donor to recipient by a direct cell to cell conjugation through conjugation tube. This conjugation tube brings physical contact between two cells.(c)In transduction, phage acts as a vehicle for gene transfer from one bacterium to another. There is no contact between donor and recipient. Virus particle takes up the DNA fragment from the donor bacterium and transfers it to another bacterium which is the recipient.
(d)Lytic cycle is a process in which virus i.e. phage particle overtakes the cell and uses its machinery to reproduce. Multiple copies of virus particles cause bursting, lysis of cell and release new virus particles to infect other cells.(e)In lysogeny, virus particles incorporate its genetic material into the genome of host i.e. integration of phage DNA into a chromosome of the host cell. This integration allows a virus to lie in the dormant state within the host until it enters the lytic phase.(f)Episome is the small circular DNA molecule which is the portion of genetic material that can exist independently of main body of chromosome or can integrate into chromosome. Insertion sequences, transposons and viruses are examples of episomes which integrate with genetic material and affect its normal functioning. Virus episomes destroy the host cells. Transposons when integrate into chromosome may provide resistance to cell.(g)Auxotroph is the mutant strain that has lost their ability to synthesize certain substances. So, they require additional nutrient supplements to those of normal organisms. These additional substances in the medium of growth required for their growth and metabolism because of mutational changes.(h)Pangenome is a sum of core genome and variable genome. Core genome is the set of all genes that are shared by all bacterial species and variable genome is set of genes which is present in some specific strains or subsets of strains. This variable genome is unique to individual strains.(i)Gene targeting is a technique which utilizes homologous recombination between engineered exogenous DNA and genome of an individual. This method mutates gene in bacterial genome by delete a gene, removing exons, adding additional gene or by any point mutation. This technique is widely used for studying genetic disease or introducing specific mutation of interest in organisms.
(d)Lytic cycle is a process in which virus i.e. phage particle overtakes the cell and uses its machinery to reproduce. Multiple copies of virus particles cause bursting, lysis of cell and release new virus particles to infect other cells.(e)In lysogeny, virus particles incorporate its genetic material into the genome of host i.e. integration of phage DNA into a chromosome of the host cell. This integration allows a virus to lie in the dormant state within the host until it enters the lytic phase.(f)Episome is the small circular DNA molecule which is the portion of genetic material that can exist independently of main body of chromosome or can integrate into chromosome. Insertion sequences, transposons and viruses are examples of episomes which integrate with genetic material and affect its normal functioning. Virus episomes destroy the host cells. Transposons when integrate into chromosome may provide resistance to cell.(g)Auxotroph is the mutant strain that has lost their ability to synthesize certain substances. So, they require additional nutrient supplements to those of normal organisms. These additional substances in the medium of growth required for their growth and metabolism because of mutational changes.(h)Pangenome is a sum of core genome and variable genome. Core genome is the set of all genes that are shared by all bacterial species and variable genome is set of genes which is present in some specific strains or subsets of strains. This variable genome is unique to individual strains.(i)Gene targeting is a technique which utilizes homologous recombination between engineered exogenous DNA and genome of an individual. This method mutates gene in bacterial genome by delete a gene, removing exons, adding additional gene or by any point mutation. This technique is widely used for studying genetic disease or introducing specific mutation of interest in organisms.
2
The unicellular, rod-shaped bacterium E. coli is 2 m long and 0.8 m wide, and has a genome consisting of a single 4.6 Mb circular DNA molecule. The unicellular archaean Methanosarcina acetivorans is spherical (coccus-shaped) with a diameter of 3 m and has a 5.7 Mb circular genome. The unicellular eukaryote Saccharomyces cerevisiae is roughly spherical, with a diameter of 5-10 m. It has a haploid genome of 12 Mb divided among 16 linear chromosomes. Given these descriptions, how could you determine whether a new, uncharacterized microorganism was a bacterium, an archaean, or a eukaryote
In the given data, there are three major criteria are given regarding 
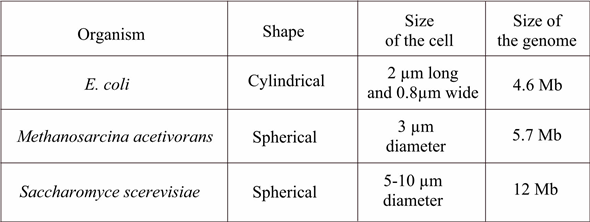 If we must determine an unknown organism based on given descriptions, then if the genome of the organism is having introns then it might be Archaean or eukaryote but not bacterium because bacterium doesn't contain any introns. Shape of the organism cannot be used as criteria for determination because it is not the perfect description of any organism to determine its class.
If we must determine an unknown organism based on given descriptions, then if the genome of the organism is having introns then it might be Archaean or eukaryote but not bacterium because bacterium doesn't contain any introns. Shape of the organism cannot be used as criteria for determination because it is not the perfect description of any organism to determine its class.
Now we must distinguish between Archaean and eukaryote then size of organism or genome size is the best way to determine between them because size of eukaryote is bigger than Archaean in both respects i.e. organism size as well as genome size. Hence in this way, we can determine the uncharacterized microorganism based on given descriptions.
 If we must determine an unknown organism based on given descriptions, then if the genome of the organism is having introns then it might be Archaean or eukaryote but not bacterium because bacterium doesn't contain any introns. Shape of the organism cannot be used as criteria for determination because it is not the perfect description of any organism to determine its class.
If we must determine an unknown organism based on given descriptions, then if the genome of the organism is having introns then it might be Archaean or eukaryote but not bacterium because bacterium doesn't contain any introns. Shape of the organism cannot be used as criteria for determination because it is not the perfect description of any organism to determine its class. Now we must distinguish between Archaean and eukaryote then size of organism or genome size is the best way to determine between them because size of eukaryote is bigger than Archaean in both respects i.e. organism size as well as genome size. Hence in this way, we can determine the uncharacterized microorganism based on given descriptions.
3
Now that the sequence of the entire E. coli K12 strain genome (roughly 5 Mb) is known, you can determine exactly where a cloned fragment of DNA came from in the genome by sequencing a few bases and matching that data with genomic information.
a. About how many nucleotides of sequence information would you need to determine exactly where a fragment is from
b. If you had purified a protein from E. coli cells, roughly how many amino acids of that protein would you need to know to establish which gene encoded the protein
c. You determine 100 nucleotides of sequence of genomic DNA from a different E. coli strain, but you cannot find a match in the E. coli K12 genome sequence. How is this possible
a. About how many nucleotides of sequence information would you need to determine exactly where a fragment is from
b. If you had purified a protein from E. coli cells, roughly how many amino acids of that protein would you need to know to establish which gene encoded the protein
c. You determine 100 nucleotides of sequence of genomic DNA from a different E. coli strain, but you cannot find a match in the E. coli K12 genome sequence. How is this possible
a.Approximately 23 nucleotides information is needed to determine the fragment's location.b.23 nucleotides would code for 7 amino acids (23/3=7.6), so 7 amino acids of the protein would be needed to establish the gene that is synthesizing the protein.c.This is possible, since Escherichia coli K12 is a genetically engineered microbe, whereas, normal E.coli is wild type strain. So, the sequence that is a part of the wild type E.coli cannot be found in the K12 strain.
4
Bacterial genomes such as that of E. coli typically have only a single origin of replication, from which replication proceeds bidirectionally. PolIII, the DNA polymerase responsible for replicating the E. coli chromosome, synthesizes DNA at a rate of about 1000 nucleotides per second.
a. From this information, estimate the minimum generation time of E. coli.
b. Under optimal conditions, E. coli have been observed to divide in as little as 17 minutes. Speculate how this might be possible, given your answer to part (a).
a. From this information, estimate the minimum generation time of E. coli.
b. Under optimal conditions, E. coli have been observed to divide in as little as 17 minutes. Speculate how this might be possible, given your answer to part (a).

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
5
List at least three features of eukaryotic genomes that are not found in bacterial genomes.

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
6
Describe a mechanism by which a gene could move from the bacterial genome to a plasmid in the same cell, or vice versa.

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
7
High salt concentrations tend to cause protein aggregation. Suggest a way to identify proteins normally expressed in particular bacterial species that can retain their solubility despite high salt conditions.

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
8
Linezolid is a new type of antibiotic that inhibits protein synthesis in several bacterial species by binding to the 50S subunit of the ribosome and inhibiting its ability to participate in the formation of translational initiation complexes. Physicians are particularly interested in this antibiotic for treating pneumonia caused by penicillin-resistant Streptococcus pneumoniae (also called ("pneumococci"). To explore the mechanisms by which pneumococci can develop resistance to linezolid, you want first to identify linezolid - resistant mutant strains. Next, using one of these strains as starting material, you now want to identify derivatives of these mutants that are no longer tolerant of linezolid.
a. Outline the techniques you would use to identify linezolid-resistant mutant pneumococci and linezolid-sensitive derivatives of these mutants. In each case, would your techniques involve direct selection, screening, replica plating, enrichment, treating with mutagens, or testing for a visible phenotype
b. Suggest possible mutations that could be responsible for the two kinds of phenotypes you will identify. What types of events in the bacterial cells would be altered by the mutations Can you classify these mutations as loss-of-function or gain- of-function
a. Outline the techniques you would use to identify linezolid-resistant mutant pneumococci and linezolid-sensitive derivatives of these mutants. In each case, would your techniques involve direct selection, screening, replica plating, enrichment, treating with mutagens, or testing for a visible phenotype
b. Suggest possible mutations that could be responsible for the two kinds of phenotypes you will identify. What types of events in the bacterial cells would be altered by the mutations Can you classify these mutations as loss-of-function or gain- of-function

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
9
A liquid culture of E. coli at a concentration of 2 × 10 8 cells/ml was diluted serially, as shown in the following diagram, and 0.1 ml of cells from the last two test tubes were spread on agar plates containing rich media. How many colonies do you expect to grow on each of the two plates 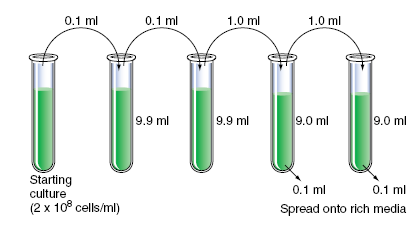


Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
10
Pick out the media (i, ii, iii, or iv) onto which you would spread cells from a Lac - Met - E. coli culture to
a. select for Lac + cells
b. screen for Lac + cells
c. select for Met + cells
i. minimal media + glucose + methionine
ii. minimal media + glucose (no methionine)
iii. rich media + X - Gal
iv. minimal media + lactose + methionine
a. select for Lac + cells
b. screen for Lac + cells
c. select for Met + cells
i. minimal media + glucose + methionine
ii. minimal media + glucose (no methionine)
iii. rich media + X - Gal
iv. minimal media + lactose + methionine

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
11
This problem concerns Fig. 13.14 on p. 471, which illustrates the experiment performed by Lederberg and Tatum that first indicated the existence of bacterial conjugation.
a. Strain A had mutations in two genes, while strain B had three mutations. The reason is that Lederberg and Tatum wanted to ensure that the phenomenon they were examining did not involve reversion of mutations. Explain the logic behind this aspect of their experimental design, assuming that the rate of reversion of a single gene is one in 10 million (1 in 10 7 ) cells. How did these investigators know that the cells they found after mixing the two cultures were indeed not due to reversion
b. The experiment shown in Fig. 13.14 did not inform the investigators which strain was the donor and which was the recipient. Describe a way in which they could modify this experiment to answer this question.
Figure 13.14 Conjugation. Neither of two multiple auxotrophic strains analyzed by Lederberg and Tatum formed colonies on minimal medium. When cells of the two strains were mixed, gene transfer produced some prototrophic cells that formed colonies on minimal medium.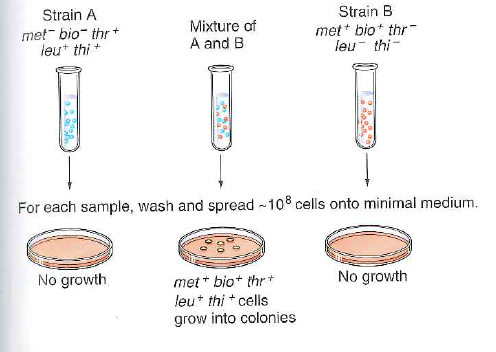
a. Strain A had mutations in two genes, while strain B had three mutations. The reason is that Lederberg and Tatum wanted to ensure that the phenomenon they were examining did not involve reversion of mutations. Explain the logic behind this aspect of their experimental design, assuming that the rate of reversion of a single gene is one in 10 million (1 in 10 7 ) cells. How did these investigators know that the cells they found after mixing the two cultures were indeed not due to reversion
b. The experiment shown in Fig. 13.14 did not inform the investigators which strain was the donor and which was the recipient. Describe a way in which they could modify this experiment to answer this question.
Figure 13.14 Conjugation. Neither of two multiple auxotrophic strains analyzed by Lederberg and Tatum formed colonies on minimal medium. When cells of the two strains were mixed, gene transfer produced some prototrophic cells that formed colonies on minimal medium.


Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
12
In two isolates (one is resistant to ampicillin and the other is sensitive to ampicillin) of a new bacterium, you found that genes encoding ampicillin resistance are being transferred into the sensitive strain.
a. How would you know that gene transfer is taking place
b. To determine if the gene transfer is transformation or transduction, you treat the mixed culture of cells with DNase. Why would this treatment distinguish between these two modes of gene transfer Describe the results predicted if the gene transfer is transformation versus transduction.
c. To determine if the gene transfer involves transformation, conjugation, or transduction, you separate the ampicillin-resistant and ampicillin-sensitive strains by a membrane with pores that are smaller than the size of a bacterium, but larger than the sizes of bacteriophage or DNA fragments. If gene transfer is still observed, what mechanisms are possibly involved and which are excluded
a. How would you know that gene transfer is taking place
b. To determine if the gene transfer is transformation or transduction, you treat the mixed culture of cells with DNase. Why would this treatment distinguish between these two modes of gene transfer Describe the results predicted if the gene transfer is transformation versus transduction.
c. To determine if the gene transfer involves transformation, conjugation, or transduction, you separate the ampicillin-resistant and ampicillin-sensitive strains by a membrane with pores that are smaller than the size of a bacterium, but larger than the sizes of bacteriophage or DNA fragments. If gene transfer is still observed, what mechanisms are possibly involved and which are excluded

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
13
E. coli cells usually have only one copy of the F plasmid per cell. You have isolated a cell in which a mutation increases the copy number of F to three to four per cell. How could you distinguish between the possibility that the copy number change was due to a mutation in the F plasmid versus a mutation in a chromosomal gene

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
14
In E. coli, the genes purC and pyrB are located halfway around the chromosome from each other. These genes are never cotransformed. Why not

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
15
DNA sequencing of the entire H. influenzae genome was completed in 1995. When DNA from the nonpathogenic strain H. influenzae Rd was compared to that of the pathogenic b strain, eight genes of the fimbrial gene cluster (located between the purE and pepN genes) involved in adhesion of bacteria to host cells were completely missing from the nonpathogenic strain. What effect would this have on cotrans formation of purE and pepN genes using DNA isolated from the nonpathogenic versus the pathogenic strain

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
16
Genes encoding toxins are often located on plasmids. There has been a recent outbreak in which a bacterium that is usually nonpathogenic is producing a toxin. Plasmid DNA can be isolated from this newly pathogenic bacterial strain and separated from the chromosomal DNA. To determine if the plasmid DNA contains a gene encoding the toxin, you could determine the sequence of the entire plasmid and search for a sequence that looks like other toxin genes previously identified. There is an easier way to determine whether the plasmid DNA carries the gene(s) for the toxin that does not involve DNA sequence analysis. Describe an experiment using this easier method.

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
17
a. You want to perform an interrupted mating map ping with an Hfr strain that is Pyr + , Met + , Xyl + , Tyr + , Arg + , His + , Mal + , and Str s. Describe an appropriate bacterial strain to be used as the other partner in this mating.
b. In an Hfr × F - cross, the pyrE gene enters the recipient in 5 minutes, but at this time point there are no exconjugants that are Met + , Xyl + , Tyr + , Arg + , His + , or Mal +. The mating is now allowed to proceed for 30 minutes and Pyr + exconjugants are selected. Of the Pyr + cells, 32% are Met + , 94% are Xyl + , 7% are Tyr + , 59% are Arg + , 0% are His + , 71% are Mal+. What can you conclude about the order of the genes
b. In an Hfr × F - cross, the pyrE gene enters the recipient in 5 minutes, but at this time point there are no exconjugants that are Met + , Xyl + , Tyr + , Arg + , His + , or Mal +. The mating is now allowed to proceed for 30 minutes and Pyr + exconjugants are selected. Of the Pyr + cells, 32% are Met + , 94% are Xyl + , 7% are Tyr + , 59% are Arg + , 0% are His + , 71% are Mal+. What can you conclude about the order of the genes

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
18
The Problem require you to diagram recombination events that can replace specific genes on the chromosome of a recipient cell with copies of those genes introduced from a donor cell. As seen in the solution to Solved Problem I, only an even number of crossovers can produce viable recombinant chromosomes. Gene mapping is simplified if you remember that progeny classes that result from four crossovers are found much less frequently than progeny classes that require two crossovers.
In Problem 17, do you think that most of the Pyr + Arg + exconjugants are also Xyl + and Mal + , or not Explain your answer by considering the recombination events that would be required to generate colonies that are Pyr + Arg + Xyl + Mai + and those required to make Pyr + Arg + Xyl Mai colonies.
Problem 17
Solved Problem I
In Problem 17, do you think that most of the Pyr + Arg + exconjugants are also Xyl + and Mal + , or not Explain your answer by considering the recombination events that would be required to generate colonies that are Pyr + Arg + Xyl + Mai + and those required to make Pyr + Arg + Xyl Mai colonies.
Problem 17

Solved Problem I


Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
19
The Problem require you to diagram recombination events that can replace specific genes on the chromosome of a recipient cell with copies of those genes introduced from a donor cell. As seen in the solution to Solved Problem I, only an even number of crossovers can produce viable recombinant chromosomes. Gene mapping is simplified if you remember that progeny classes that result from four crossovers are found much less frequently than progeny classes that require two crossovers.
One issue with interrupted-mating experiments such as that in Problem 17 is that gene order may be ambiguous if the genes are close together. Another shortcoming is that such experiments do not provide accurate map distances. The reason is that researchers select for the first Hfr marker transferred into the recipient, but the recovery of F exconjugants with a later Hfr marker is complex, depending both on transfer of the marker into the cell and on crossovers that transfer the marker into the recipient chromosome.
To make more accurate maps, bacterial geneticists often do Hfr × F crosses in a different way: They select for exconjugants that contain a late Hfr marker, and then screen for the presence of the earlier markers. This method ensures that all of the markers have entered the F cell, so relative gene distances now solely reflect crossover frequencies. Furthermore, gene order is clarified by considering the crossovers responsible for each class of exconjugants.
As an example, suppose you performed the same cross as in Problem 17, but you selected for Arg 1 exconjugants, and then screened them for the earlier Hfr markers Mal + Xyl + and Pyr +. You obtained the following data: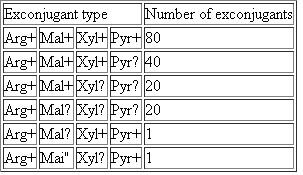 a. Explain why four of the exconjugant types are much more frequent than the other two.
a. Explain why four of the exconjugant types are much more frequent than the other two.
b. What can you conclude about the relative distances between the four genes
c. The data allow you to estimate one other relevant genetic distance. Explain.
Problem 17
Solved Problem I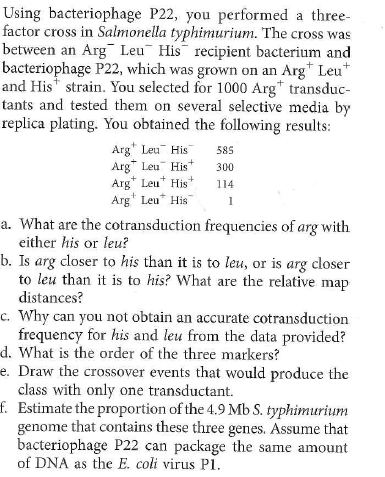
One issue with interrupted-mating experiments such as that in Problem 17 is that gene order may be ambiguous if the genes are close together. Another shortcoming is that such experiments do not provide accurate map distances. The reason is that researchers select for the first Hfr marker transferred into the recipient, but the recovery of F exconjugants with a later Hfr marker is complex, depending both on transfer of the marker into the cell and on crossovers that transfer the marker into the recipient chromosome.
To make more accurate maps, bacterial geneticists often do Hfr × F crosses in a different way: They select for exconjugants that contain a late Hfr marker, and then screen for the presence of the earlier markers. This method ensures that all of the markers have entered the F cell, so relative gene distances now solely reflect crossover frequencies. Furthermore, gene order is clarified by considering the crossovers responsible for each class of exconjugants.
As an example, suppose you performed the same cross as in Problem 17, but you selected for Arg 1 exconjugants, and then screened them for the earlier Hfr markers Mal + Xyl + and Pyr +. You obtained the following data:
 a. Explain why four of the exconjugant types are much more frequent than the other two.
a. Explain why four of the exconjugant types are much more frequent than the other two.b. What can you conclude about the relative distances between the four genes
c. The data allow you to estimate one other relevant genetic distance. Explain.
Problem 17

Solved Problem I


Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
20
Suppose you have two Hfr strains of E. coli (HfrA and HfrB), derived from a fully prototrophic streptomycin- sensitive (wild-type) F + strain. In separate experiments you allow these two Hfr strains to conjugate with an F - recipient strain (Rcp) that is streptomycin resistant and auxotrophic for glycine (Gly - ), lysine (Lys - ), nicotinic acid (Nic - ), phenylalanine (Phe - ), tyrosine (Tyr - ). and uracil (Ura - ). By using an interrupted mating protocol you determined the earliest time after mating at which each of the markers can be detected in the streptomycin-resistant recipient strain, as shown here. 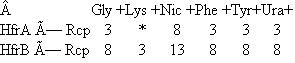 (The * indicates that no Lys + cells were recovered in the 60 minutes of the experiment.)
(The * indicates that no Lys + cells were recovered in the 60 minutes of the experiment.)
a. Draw the best map you can from these data, showing the relative locations of the markers and the origins of transfer in strains HfrA and HfrB. Show distances where possible.
b. To resolve ambiguities in the preceding map, you studied cotransduction of the markers by the generalized transducing phage P1. You grew phage P1 on strain HfrB and then used the lysate to infect strain Rcp. You selected 1000 Phe + clones and tested them for the presence of unselected markers, with the following results: Draw the order of the genes as best you can based on the preceding cotransduction data. c. Suppose you wanted to use generalized transduction to map the gly gene relative to at least some of the other markers. How would you modify the co- transduction experiment just described to increase your chances of success Describe the composition of the medium you would use.
Draw the order of the genes as best you can based on the preceding cotransduction data. c. Suppose you wanted to use generalized transduction to map the gly gene relative to at least some of the other markers. How would you modify the co- transduction experiment just described to increase your chances of success Describe the composition of the medium you would use.
 (The * indicates that no Lys + cells were recovered in the 60 minutes of the experiment.)
(The * indicates that no Lys + cells were recovered in the 60 minutes of the experiment.)a. Draw the best map you can from these data, showing the relative locations of the markers and the origins of transfer in strains HfrA and HfrB. Show distances where possible.
b. To resolve ambiguities in the preceding map, you studied cotransduction of the markers by the generalized transducing phage P1. You grew phage P1 on strain HfrB and then used the lysate to infect strain Rcp. You selected 1000 Phe + clones and tested them for the presence of unselected markers, with the following results:
 Draw the order of the genes as best you can based on the preceding cotransduction data. c. Suppose you wanted to use generalized transduction to map the gly gene relative to at least some of the other markers. How would you modify the co- transduction experiment just described to increase your chances of success Describe the composition of the medium you would use.
Draw the order of the genes as best you can based on the preceding cotransduction data. c. Suppose you wanted to use generalized transduction to map the gly gene relative to at least some of the other markers. How would you modify the co- transduction experiment just described to increase your chances of success Describe the composition of the medium you would use.
Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
21
The Problem require you to diagram recombination events that can replace specific genes on the chromosome of a recipient cell with copies of those genes introduced from a donor cell. As seen in the solution to Solved Problem I, only an even number of crossovers can produce viable recombinant chromosomes. Gene mapping is simplified if you remember that progeny classes that result from four crossovers are found much less frequently than progeny classes that require two crossovers.
Starting with an F strain that was prototrophic (that is, had no auxotrophic mutations) and Str S , several independent Hfr strains were isolated. These Hfr strains were mated to an F + strain that was Str r Arg Cys His Ilv Lys Met Nic Pab Pyr Trp . Interrupted-mating experiments showed that the Hfr strains transferred the wild-type alleles in the order listed in the following table as a function of time. The time of entry for the markers within parentheses could not be distinguished from one another. a. From these data, derive a map of the relative position of these markers on the bacterial chromosome. Indicate with labeled arrows the position and orientation of the integrated F plasmid for each Hfr strain.
a. From these data, derive a map of the relative position of these markers on the bacterial chromosome. Indicate with labeled arrows the position and orientation of the integrated F plasmid for each Hfr strain.
b. To determine the relative order of the trp , pyr , and cys markers and the distances between them, HfrB was mated with the F strain long enough to allow transfer of the nic marker, after which Trp + recombinants were selected. The unselected markers pyr and cys were then scored in the Trp + recombinants, yielding the following results: Draw a map of the trp, pyr, and cys markers relative to each other. (Note that you cannot determine the order relative to the nic or his genes using these data.) Express map distances between adjacent genes as the frequency of crossing-over between them.
Draw a map of the trp, pyr, and cys markers relative to each other. (Note that you cannot determine the order relative to the nic or his genes using these data.) Express map distances between adjacent genes as the frequency of crossing-over between them.
Solved Problem I
Starting with an F strain that was prototrophic (that is, had no auxotrophic mutations) and Str S , several independent Hfr strains were isolated. These Hfr strains were mated to an F + strain that was Str r Arg Cys His Ilv Lys Met Nic Pab Pyr Trp . Interrupted-mating experiments showed that the Hfr strains transferred the wild-type alleles in the order listed in the following table as a function of time. The time of entry for the markers within parentheses could not be distinguished from one another.
 a. From these data, derive a map of the relative position of these markers on the bacterial chromosome. Indicate with labeled arrows the position and orientation of the integrated F plasmid for each Hfr strain.
a. From these data, derive a map of the relative position of these markers on the bacterial chromosome. Indicate with labeled arrows the position and orientation of the integrated F plasmid for each Hfr strain.b. To determine the relative order of the trp , pyr , and cys markers and the distances between them, HfrB was mated with the F strain long enough to allow transfer of the nic marker, after which Trp + recombinants were selected. The unselected markers pyr and cys were then scored in the Trp + recombinants, yielding the following results:
 Draw a map of the trp, pyr, and cys markers relative to each other. (Note that you cannot determine the order relative to the nic or his genes using these data.) Express map distances between adjacent genes as the frequency of crossing-over between them.
Draw a map of the trp, pyr, and cys markers relative to each other. (Note that you cannot determine the order relative to the nic or his genes using these data.) Express map distances between adjacent genes as the frequency of crossing-over between them.Solved Problem I


Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
22
You can carry out matings between an Hfr and F - strain by mixing the two cell types in a small patch on a plate and then replica plating to selective media. This methodology was used to screen hundreds of different cells for a recombination-deficient recA - mutant. Why is this an assay for RecA function Would you be screening for a recA - mutation in the F - or Hfr strain using this protocol

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
23
Genome sequences show that some pathogenic bacteria contain virulence genes next to bacteriophage genes. Why does this suggest lateral gene transfer, and what would the mechanism of transfer have been

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
24
Generalized and specialized transduction both involve bacteriophages. What are the differences between these two types of transduction

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
25
This problem highlights some useful variations of the gene identification by plasmid transformation procedure shown in Fig. 13.28 on p. 480.
a. Suppose you have obtained a new bacterial mutant strain of interest. To determine the affected gene, you sequence the entire genome of the mutant strain and compare it with that of a wild-type strain. Several differences between the genomes were found, but one of these is a nonsense mutation that seems to be a good candidate. How would you use a plasmid library to verify that this nonsense mutation is responsible for the mutant phenotype
b. Figure 13.28 showed how plasmid libraries could be used to identify genes with loss-of-function mutations that are responsible for a given aberrant phenotype. How could you use a plasmid library to identify a gene affected by a gain-of-function mutation
Figure 13.28 Identifying mutant genes by plasmid library transformation. In this example, mutant auxotrophic bacterial cells ( arg ) are transformed with a recombinant library made from wild-type E. coli genomic DNA. The plasmid purified from a colony that grows on minimal media without an arginine supplement contains the arg + allele of the gene that is mutant in the auxotrophs.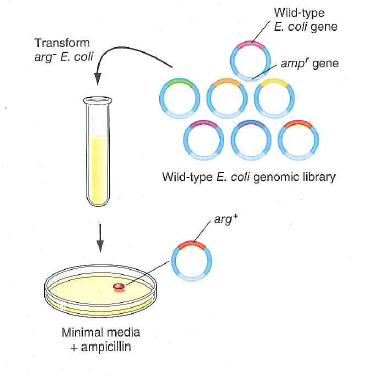
a. Suppose you have obtained a new bacterial mutant strain of interest. To determine the affected gene, you sequence the entire genome of the mutant strain and compare it with that of a wild-type strain. Several differences between the genomes were found, but one of these is a nonsense mutation that seems to be a good candidate. How would you use a plasmid library to verify that this nonsense mutation is responsible for the mutant phenotype
b. Figure 13.28 showed how plasmid libraries could be used to identify genes with loss-of-function mutations that are responsible for a given aberrant phenotype. How could you use a plasmid library to identify a gene affected by a gain-of-function mutation
Figure 13.28 Identifying mutant genes by plasmid library transformation. In this example, mutant auxotrophic bacterial cells ( arg ) are transformed with a recombinant library made from wild-type E. coli genomic DNA. The plasmid purified from a colony that grows on minimal media without an arginine supplement contains the arg + allele of the gene that is mutant in the auxotrophs.


Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
26
A researcher has a trp auxotrophic strain of E. coli with a mutation in a single gene. To identify that mutant gene, she uses a genomic library made from a wild-type version of that same strain to find plasmids that rescue the mutant phenotype. The result is surprising. She recovers 10 plasmids that provide a Trp+ phenotype, but six of the plasmids contain gene X, while the other four contain gene Y. Our scientist has encountered a phenomenon called multicopy suppression related to the fact that plasmids are usually present in several copies per bacterium. Because the genes in the plasmids are present in more than their usual single copy in the bacterial chromosome, more than the usual amount of Protein X or Protein Y is being produced from the plasmids. Sometimes, overexpression of one protein can rescue the mutant phenotype caused by loss of a different protein. Suggest at least two ways that our scientist could determine which of the two genes, gene X or gene Y, actually corresponds to the mutant gene causing the Trp phenotype

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
27
Streptococcus parasanguis is a bacterial species that initiates dental plaque formation by adhering to teeth. To investigate ways to eliminate plaque, researchers constructed a plasmid, depicted in the figure shown, to mutagenize S. parasanguis. The key features of this plasmid include repA ts (a temperature-sensitive origin of replication), kan r (a gene for resistance to the antibiotic kanamycin), and the transposon 1S256. This transposon contains the erm r gene for resistance to the antibiotic erythromycin and transposes in S. parasanguis thanks to a gene encoding a transposase enzyme that moves all DNA sequences located between the transposons inverted repeats [IRs]. ![Streptococcus parasanguis is a bacterial species that initiates dental plaque formation by adhering to teeth. To investigate ways to eliminate plaque, researchers constructed a plasmid, depicted in the figure shown, to mutagenize S. parasanguis. The key features of this plasmid include repA ts (a temperature-sensitive origin of replication), kan r (a gene for resistance to the antibiotic kanamycin), and the transposon 1S256. This transposon contains the erm r gene for resistance to the antibiotic erythromycin and transposes in S. parasanguis thanks to a gene encoding a transposase enzyme that moves all DNA sequences located between the transposons inverted repeats [IRs]. a. How could the researchers use this plasmid as a mutagen Consider how they could get the transposon into the bacteria, and how they could identify strains that had new insertions of IS256 into S. parasanguis genes. Your answer should explain why the plasmid has two different antibiotic resistance genes as well as a temperature-sensitive origin of replication. b. Why would the researchers use this plasmid as a mutagen c. If the investigators found a mutant strain of S. parasanguis that was defective in plaque formation, how could they identify the affected gene](https://storage.examlex.com/SM1277/11eb58c5_c2ed_d49d_8125_65e18d37b7c9_SM1277_00.jpg)
a. How could the researchers use this plasmid as a mutagen Consider how they could get the transposon into the bacteria, and how they could identify strains that had new insertions of IS256 into S. parasanguis genes. Your answer should explain why the plasmid has two different antibiotic resistance genes as well as a temperature-sensitive origin of replication.
b. Why would the researchers use this plasmid as a mutagen
c. If the investigators found a mutant strain of S. parasanguis that was defective in plaque formation, how could they identify the affected gene
![Streptococcus parasanguis is a bacterial species that initiates dental plaque formation by adhering to teeth. To investigate ways to eliminate plaque, researchers constructed a plasmid, depicted in the figure shown, to mutagenize S. parasanguis. The key features of this plasmid include repA ts (a temperature-sensitive origin of replication), kan r (a gene for resistance to the antibiotic kanamycin), and the transposon 1S256. This transposon contains the erm r gene for resistance to the antibiotic erythromycin and transposes in S. parasanguis thanks to a gene encoding a transposase enzyme that moves all DNA sequences located between the transposons inverted repeats [IRs]. a. How could the researchers use this plasmid as a mutagen Consider how they could get the transposon into the bacteria, and how they could identify strains that had new insertions of IS256 into S. parasanguis genes. Your answer should explain why the plasmid has two different antibiotic resistance genes as well as a temperature-sensitive origin of replication. b. Why would the researchers use this plasmid as a mutagen c. If the investigators found a mutant strain of S. parasanguis that was defective in plaque formation, how could they identify the affected gene](https://storage.examlex.com/SM1277/11eb58c5_c2ed_d49d_8125_65e18d37b7c9_SM1277_00.jpg)
a. How could the researchers use this plasmid as a mutagen Consider how they could get the transposon into the bacteria, and how they could identify strains that had new insertions of IS256 into S. parasanguis genes. Your answer should explain why the plasmid has two different antibiotic resistance genes as well as a temperature-sensitive origin of replication.
b. Why would the researchers use this plasmid as a mutagen
c. If the investigators found a mutant strain of S. parasanguis that was defective in plaque formation, how could they identify the affected gene

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
28
The sequence at one end of one strand of the Drosophila transposon Mariner is shown below (dots indicate sequences within the transposon): 
You obtain a mutant bacterial strain tagged with an engineered Mariner transposon, cut the genomic DNA from this strain with the restriction enzyme Mbo I(whose recognition site is ^GATC), and circularize the resultant DNA fragments by diluting the restriction enzyme digest and adding DNA ligase.
a. Design two 17 bp PCR primers that you could use to identify (by inverse PCR) the gene into which the transposon inserted. Your primers should amplify the smallest possible DNA fragment whose sequence would allow this determination.
b. What DNA sequence will be amplified from the circularized fragments of the mutant genome Show the extent of this DNA sequence on a map of the genome of the mutant strain, indicating the locations of the transposon insertion and any relevant sites for the enzyme Mbo I.

You obtain a mutant bacterial strain tagged with an engineered Mariner transposon, cut the genomic DNA from this strain with the restriction enzyme Mbo I(whose recognition site is ^GATC), and circularize the resultant DNA fragments by diluting the restriction enzyme digest and adding DNA ligase.
a. Design two 17 bp PCR primers that you could use to identify (by inverse PCR) the gene into which the transposon inserted. Your primers should amplify the smallest possible DNA fragment whose sequence would allow this determination.
b. What DNA sequence will be amplified from the circularized fragments of the mutant genome Show the extent of this DNA sequence on a map of the genome of the mutant strain, indicating the locations of the transposon insertion and any relevant sites for the enzyme Mbo I.

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
29
Scientists can use gene targeting not just to knock genes out (as was shown in Fig. 13.31 on p. 481), but also to introduce nonbacterial genes into bacterial chromosomes. One such gene in wide use is a gene from jellyfish encoding Green Fluorescent Protein. In one example of this strategy, suppose you want to make E. coli into a biosensor to detect the highly toxic metal cadmium. The E. coli genome has a gene called yodA that is only transcribed (and its mRNA translated) in the presence of cadmium. You want to use gene targeting to make a strain of E. coli that will fluoresce brightly in green when cadmium is present in the environment.
a. Draw a DNA construct that you could use to exchange the yodA coding sequence with that for jellyfish Green Fluorescent Protein. Would you obtain the jellyfish DNA from genomic DNA or from a cDNA clone
b. Explain why yodA is no longer functional in bacteria that glow green in the presence of cadmium.
c. Can you think of way to alter the approach so that yodA might remain functional
Figure 13.31 Gene targeting. Linear DNA fragments (generated using PCR) introduced into E. coli undergo recombination with bacterial chromosome sequences homologous to the fragment ' s free ends. Here, Incorporation of the DNA fragment replaces most of gene X with an ampicillin resistance gene , generating gene X null mutants. Cells that have undergone gene X replacement can be selected by growing on medium containing ampicillin.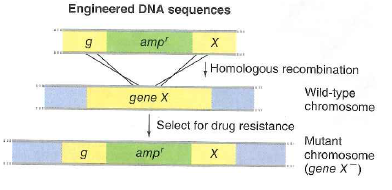
a. Draw a DNA construct that you could use to exchange the yodA coding sequence with that for jellyfish Green Fluorescent Protein. Would you obtain the jellyfish DNA from genomic DNA or from a cDNA clone
b. Explain why yodA is no longer functional in bacteria that glow green in the presence of cadmium.
c. Can you think of way to alter the approach so that yodA might remain functional
Figure 13.31 Gene targeting. Linear DNA fragments (generated using PCR) introduced into E. coli undergo recombination with bacterial chromosome sequences homologous to the fragment ' s free ends. Here, Incorporation of the DNA fragment replaces most of gene X with an ampicillin resistance gene , generating gene X null mutants. Cells that have undergone gene X replacement can be selected by growing on medium containing ampicillin.


Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
30
Scientists who study amino acid biosynthesis pathways want to isolate auxotrophic bacteria. A technique called penicillin enrichment makes this task easier. This procedure starts by exposing a liquid culture of wild-type (prototrophic) bacteria growing in rich (complete) medium to a chemical mutagen. After this treatment, the cells are centrifuged to remove the liquid and the mutagen. The pellet of cells at the bottom of the centrifuge tube is now resuspended in medium that lacks one amino acid (in this example, cysteine) but contains penicillin. Subsequently, the bacteria are poured onto a filter that concentrates them and allows them to be washed free of the penicillin. The living, bacteria retained on the filter are highly enriched for cysteine auxotrophs.
a. Given what you know about the action of pencillin, explain why this enrichment occurs.
b. Penicillin enrichment is not a selection, because the drug does not kill 100% of the prototrophs. The cells on the filter thus need to be screened for cysteine auxotrophy. How would the scientists perform this screen
c. If the starting strain contained a pen r gene on a plasmid, would this scheme still enrich for auxotrophs Explain.
a. Given what you know about the action of pencillin, explain why this enrichment occurs.
b. Penicillin enrichment is not a selection, because the drug does not kill 100% of the prototrophs. The cells on the filter thus need to be screened for cysteine auxotrophy. How would the scientists perform this screen
c. If the starting strain contained a pen r gene on a plasmid, would this scheme still enrich for auxotrophs Explain.

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
31
List key features of prokaryotic cells.

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
32
Discuss how bacterial habitats influence bacterial metabolisms.

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
33
Summarize the properties that make certain bacteria pathogenic to humans.

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
34
Describe how genes are organized within a bacterial genome.

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
35
Differentiate between a specie ' s core genome and its pangenome.

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
36
Discriminate between IS and Tn transposable elements in bacteria.

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
37
Describe how plasmids conferring multidrug resistance to bacteria may have evolved.

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
38
Explain how the study of metagenomics might yield practical benefits.

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
39
Describe the features of bacteria, particularly E. coli , that make them useful laboratory organisms.

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
40
Describe different classes of bacterial mutants and the methods for identifying them.

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
41
Compare the three mechanisms of gene transfer in bacteria: transformation, conjugation, and transduction.

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
42
Explain how each of these three methods of gene transfer can be used to map bacterial genes.

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
43
Discuss the role of horizontal gene transfer in the evolution of bacteria.

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
44
Explain how to identify mutant genes by transformation with recombinant plasmids.

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
45
Discuss the use of transposons as mutagens in bacteria.

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
46
Describe how to generate specific mutations in any E. coli gene by gene targeting.

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
47
Explain how penicillin kills bacteria.

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
48
Describe mechanisms of penicillin resistance and how N. gonorrhoeae has become resistant.

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck
49
Discuss potential solutions to the worldwide problem of drug-resistant pathogens.

Unlock Deck
Unlock for access to all 49 flashcards in this deck.
Unlock Deck
k this deck


