Exam 7: Genetic Variation Producing Diseasecausing Abnormalities in Dna and Chromosomes
Depending on the base position within as codon, the percentage of base changes that alter the interpretation of the codon vary remarkably. Which of the following statements, if any, is true?
a) 100% of all possible base changes to the first base cause an altered interpretation for the codon.
b) 100% of all possible changes to the second base cause an altered interpretation for the codon.
c) About 30% of all possible changes to the third base cause an altered interpretation for the codon.
d) Less than 10% of all possible changes to the third base cause an altered interpretation for the codon.
b) 100% of all possible changes to the second base cause an altered interpretation for the codon.
c) About 30% of all possible changes to the third base cause an altered interpretation for the codon.
A chromosome has received two double-stranded breaks. What kinds of chromosome abnormalities can result?
If the two breaks occur on the same chromosome arm, there are two possibilities. First an interstitial deletion can result, after the central piece is excised and the two ends are fused at the breakpoints (the excised central fragment cannot be propagated because it lacks a centromere). Alternatively, a paracentric inversion results (the central piece, lacking a centromere, rotates and then is fused to the end pieces).
If the two breaks occur on different chromosome arms, deletion is not normally viable: the central fragment cannot be excised and lost as it contains the centromere. Instead, there are two viable alternatives. First, a pericentric inversion results (the central piece, containing a centromere, rotates and then is fused to the end pieces). More rarely, a ring chromosome can occur where the two ends of the central fragment fuse to form a circular chromosome.
What are amyloid diseases? In what respects do neurodegenerative amyloid diseases resemble prion diseases?
Amyloid protein is the term used to describe a broad family of proteins that have a high content of -sheets that make them prone to aggregation. Prions are a well-known subclass of amyloid proteins but in addition there are many other examples. Amyloid diseases are disorders in which amyloid proteins are implicated. The aggregation of amyloid proteins can occur outside of cells (such as in the case of prion proteins and -amyloid), within nuclei (huntingtin), or within the cytoplasm (SOD1; Tau; and synuclein).
Some common amyloid diseases are not associated with neurodegeneration, such as type 2 diabetes (aggregates of serum amyloid A protein are found in the pancreatic islets of Langerhans). However, neurodegeneration is the most striking clinical characteristic of many amyloid diseases.
Alzheimer disease, Parkison disease, amyotrophic lateral sclerosis, and frontotemporal disease, resemble prion protein diseases in many ways and are sometimes classified as prionoid diseases. In these diseases direct involvement of the aggregated proteins in disease is supported from familial forms of these disorders in which mutations in the relevant gene promote the formation of amyloid protein.
There is no evidence from animal studies that the aggregated proteins in prionoid disorders are infectious like prion proteins. But there is quite strong evidence that the pathogenesis resembles prion protein disease in two respects. First, like prion proteins, misfolded amyloid proteins in these disorders can induce the formation of the amyloid state in the normal proteins so that they aggregate. Secondly, for several of the disorders there is strong evidence for cell-to-cell spreading of the disorder.
Match each of the genetic mechanisms
a) to
d) with one or more of the possible outcomes i) to iv).

Nonsynonymous mutations can be grouped into three classes. What are they?
List three examples of single gene disorders that show an extremely limited range of point mutations and explain why there should be such mutational homogeneity.
How does just a single loss-of-function allele cause a dominantly inherited disorder?
Regarding chromosome abnormalities, which of the following statements, if any, is false?
What is a cryptic splice site? What are expected consequences of activation of i) a cryptic splice donor site located within an exon ii) a cryptic splice acceptor site within an intron?
In the sequence below, the blue sequence represents an exon containing coding DNA near the beginning of a large gene and green lines and letters are flanking intron sequence. Nine mutations are shown: an insertion of an Alu repeat insertion plus three deletions at top and five single nucleotide substitutions below. Comment on the mutation class in each case and on its likely effect. 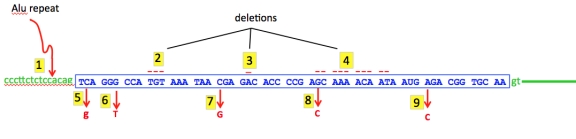
The disease mechanism in prion protein diseases has sometimes been considered a type of epigenetic mechanism. On what basis?
Certain sequence classes in our genome are particularly prone to mutations. List three examples and explain why they are so prone to mutations.
Deletions, duplications and inversions in our DNA often result in interaction between tandem repeats, direct repeats, or inverted repeats. What are the essential differences between these repeat classes.
Sequence exchange between two non-allelic copies of the same long sequence on chromosomal DNA molecules can have different consequences, depending on the positioning of the repeats that participate in sequence exchange. In (i) to (iii) imagine that there is sequence exchange between non-allelic sequence copies A and B shown by the blue arrows - the sequence exchange is between different chromatids of the same chromosome in the case of (ii). What types of sequence exchange might occur and what would be the likely outcome? 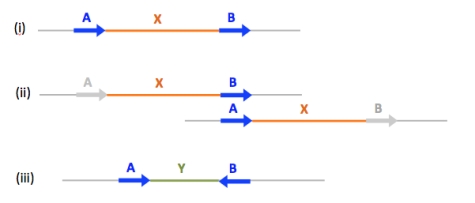
Constitutional aneuploidy is occasionally viable in humans. Why should having fewer or extra copies of certain chromosomes be compatible with life, but not so in the case of other chromosomes?
Which of the following changes is i) a synonymous mutation, ii) a conservative substitution, iii) a nonconservative substitution, iv) a stop-loss mutation, v) a stop-gain mutation.
a) UGA UCA
b) UAC UAA
c) AGA AAA
d) AGA CGA
e) UGU CGU
Why should our mitochondrial genetic code be different from our nuclear genetic code?
Match each of the characteristics in
a) to
e) with the mutant alleles listed in (i) to (v).

Studies of single gene disorders have sought to draw correlations between the genotypes at a disease locus and the phenotype of the single gene but the genotype-phenotype correlations are often poor. Even within families there may be significant variability in the phenotype of affected members (who are expected or known to have the same genotypes at the disease locus). List three factors that can explain why that should be so.
Filters
- Essay(0)
- Multiple Choice(0)
- Short Answer(0)
- True False(0)
- Matching(0)