Deck 9: Digital Analysis of Dna
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/29
Play
Full screen (f)
Deck 9: Digital Analysis of Dna
1
The mouse genome consists of 2,700 Mb. A genomic library is constructed using BAC vectors and overlapping fragments of the mouse genome. The average size of the mouse DNA inserts is 200 kb.
-How many clones would make up one genomic equivalent?
A)135
B)270
C)13,500
D)270,000
-How many clones would make up one genomic equivalent?
A)135
B)270
C)13,500
D)270,000
13,500
2
Which is a characteristic of a plasmid used as a cloning vector? (Select all that apply. )
A)contains a selectable marker
B)contains an origin of replication
C)has several unique recognition sites for restriction enzymes
D)integrates into the host chromosome
E)can hold an insert of up to 300 kb
A)contains a selectable marker
B)contains an origin of replication
C)has several unique recognition sites for restriction enzymes
D)integrates into the host chromosome
E)can hold an insert of up to 300 kb
A, B, C
3
In Sanger sequencing, what causes DNA synthesis to terminate at a specific base?
A)Restriction endonucleases cleave DNA after particular bases.
B)Fluorescent molecules produce high-energy light that inactivates DNA polymerase.
C)Incorporation of a deoxynucleotide that lacks a base causes DNA polymerase to stall.
D)Incorporation of a dideoxynucleotide that lacks a 3′ hydroxyl.
E)Incorporation of a ribonucleotide that lacks a 5' phosphate.
A)Restriction endonucleases cleave DNA after particular bases.
B)Fluorescent molecules produce high-energy light that inactivates DNA polymerase.
C)Incorporation of a deoxynucleotide that lacks a base causes DNA polymerase to stall.
D)Incorporation of a dideoxynucleotide that lacks a 3′ hydroxyl.
E)Incorporation of a ribonucleotide that lacks a 5' phosphate.
D
4
What is the purpose of ethidium bromide in DNA electrophoresis?
A)To tag DNA fragments so they can be viewed under UV light
B)To introduce mutations into the DNA
C)To fragment the DNA before separating the pieces by size
D)To join fragments of DNA together
A)To tag DNA fragments so they can be viewed under UV light
B)To introduce mutations into the DNA
C)To fragment the DNA before separating the pieces by size
D)To join fragments of DNA together

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
5
In Sanger sequencing, the role of the DNA polymerase is to
A)synthesize new DNA that is complementary to the template strand.
B)proofread base pairs and correct errors.
C)synthesize a primer in the 5'→3' direction.
D)attach DNA fragments together with covalent bonds.
A)synthesize new DNA that is complementary to the template strand.
B)proofread base pairs and correct errors.
C)synthesize a primer in the 5'→3' direction.
D)attach DNA fragments together with covalent bonds.

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
6
The mouse genome consists of 2,700 Mb. A genomic library is constructed using BAC vectors and overlapping fragments of the mouse genome. The average size of the mouse DNA inserts is 200 kb.
-What is the minimum number of clones that should be in this mouse genomic library to have a 95% chance that it contains the entire genome?
A)2,700
B)13,500
C)54,000
D)135,000
-What is the minimum number of clones that should be in this mouse genomic library to have a 95% chance that it contains the entire genome?
A)2,700
B)13,500
C)54,000
D)135,000

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
7
The mouse genome consists of 2,700 Mb. A genomic library is constructed using BAC vectors and overlapping fragments of the mouse genome. The average size of the mouse DNA inserts is 200 kb.
-A mouse genomic library is constructed so that the number of clones equals one genomic equivalent.Is it likely that this library contains the entire mouse genome?
A)No; by chance some parts of the genome will be represented more than once and others not at all.
B)Yes; that is a sufficient number clones for every part of the genome to be represented one time.
C)No; the restriction enzyme used to fragment the genome will delete some DNA on the ends of clones.
-A mouse genomic library is constructed so that the number of clones equals one genomic equivalent.Is it likely that this library contains the entire mouse genome?
A)No; by chance some parts of the genome will be represented more than once and others not at all.
B)Yes; that is a sufficient number clones for every part of the genome to be represented one time.
C)No; the restriction enzyme used to fragment the genome will delete some DNA on the ends of clones.

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
8
You performed a Sanger sequencing reaction and obtained the following read.In this figure, A = green, C = purple, G = black, T = red.The height of the peaks is unimportant.The smallest molecule is at the left of the trace. 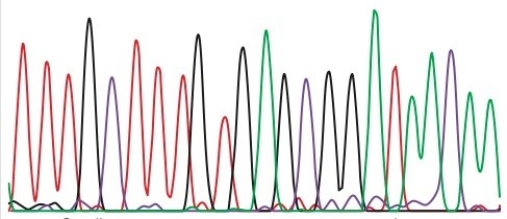
What is the sequence of the template DNA used for this sequencing reaction?
A)5' TTTGCTTTGTGAGCGGATAACAA 3'
B)3' TTTGCTTTGTGAGCGGATAACAA 5'
C)5' AAACGAAACACTCGCCTATTGTT 3'
D)3' AAACGAAACACTCGCCTATTGTT 5'

What is the sequence of the template DNA used for this sequencing reaction?
A)5' TTTGCTTTGTGAGCGGATAACAA 3'
B)3' TTTGCTTTGTGAGCGGATAACAA 5'
C)5' AAACGAAACACTCGCCTATTGTT 3'
D)3' AAACGAAACACTCGCCTATTGTT 5'

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
9
Which is an example of a recombinant DNA molecule?
A)a single-stranded RNA hybridized to a single-stranded DNA
B)a genomic fragment of human DNA ligated into a bacterial plasmid vector
C)a bacteriophage chromosome in a bacterial cell
D)a bacterial plasmid cut with a restriction enzyme and then resealed using ligase
A)a single-stranded RNA hybridized to a single-stranded DNA
B)a genomic fragment of human DNA ligated into a bacterial plasmid vector
C)a bacteriophage chromosome in a bacterial cell
D)a bacterial plasmid cut with a restriction enzyme and then resealed using ligase

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
10
You performed a Sanger sequencing reaction and obtained the following read.In this figure, A = green, C = purple, G = black, T = red.The height of the peaks is unimportant.The smallest molecule is at the left of the trace. 
What is the sequence of the DNA synthesized in the sequencing reaction?
A)5' TTTGCTTTGTGAGCGGATAACAA 3'
B)3' TTTGCTTTGTGAGCGGATAACAA 5'
C)5' AAACGAAACACTCGCCTATTGTT 3'
D)3' AAACGAAACACTCGCCTATTGTT 5'

What is the sequence of the DNA synthesized in the sequencing reaction?
A)5' TTTGCTTTGTGAGCGGATAACAA 3'
B)3' TTTGCTTTGTGAGCGGATAACAA 5'
C)5' AAACGAAACACTCGCCTATTGTT 3'
D)3' AAACGAAACACTCGCCTATTGTT 5'

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
11
You performed a Sanger sequencing reaction and obtained the following read.In this figure, A = green, C = purple, G = black, T = red.The height of the peaks is unimportant.The smallest molecule is at the left of the trace. 
Which is a step in hierarchical genome sequencing that is not found in shotgun genome sequencing?
A)fragment the genome
B)construct a genomic library
C)organize the clones based on their order in the genome
D)sequence the ends of millions of clones
E)assemble the sequences into contigs based on overlap

Which is a step in hierarchical genome sequencing that is not found in shotgun genome sequencing?
A)fragment the genome
B)construct a genomic library
C)organize the clones based on their order in the genome
D)sequence the ends of millions of clones
E)assemble the sequences into contigs based on overlap

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
12
Why isn't bacterial DNA digested by the restriction enzymes bacteria produce?
A)The sequence recognized and cleaved by a restriction enzyme is not found in the bacterial genome.
B)Bacterial cells have high concentrations of ligase which immediately repairs the any sites that are cleaved.
C)Methyl groups added to the bases of restriction enzyme recognition sites within the bacterial chromosome block cleavage.
D)Bacterial DNA is modified with phosphate groups that prevent the function of restriction enzymes.
E)Restriction enzymes are found in the cytoplasm and DNA is only in the nucleus.
A)The sequence recognized and cleaved by a restriction enzyme is not found in the bacterial genome.
B)Bacterial cells have high concentrations of ligase which immediately repairs the any sites that are cleaved.
C)Methyl groups added to the bases of restriction enzyme recognition sites within the bacterial chromosome block cleavage.
D)Bacterial DNA is modified with phosphate groups that prevent the function of restriction enzymes.
E)Restriction enzymes are found in the cytoplasm and DNA is only in the nucleus.

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
13
In gel electrophoresis, DNA fragments move at different rates because they have different
A)electrical charge densities.
B)sizes.
C)base sequences.
D)amino acid compositions.
E)overall electrical charges.
A)electrical charge densities.
B)sizes.
C)base sequences.
D)amino acid compositions.
E)overall electrical charges.

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
14
Imagine that DNA is composed of equal proportions of six different types of nucleotide, instead of the normal four.The six nucleotides are A, C, G, T, X, and Y.In this new type of DNA molecule, A pairs with T, C pairs with G, and X pairs with Y.What would be the average size of the DNA fragments produced when this new DNA is cut by a restriction endonuclease with a four-base recognition sequence?
A)125 bp
B)256 bp
C)425 bp
D)1056 bp
E)1296 bp
A)125 bp
B)256 bp
C)425 bp
D)1056 bp
E)1296 bp

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
15
What is the function of the ampr gene in a plasmid vector?
A)It provides several unique restriction enzyme recognition sites for opening up the plasmid.
B)It allows a researcher to separate bacterial cells with a plasmid from those without a plasmid.
C)It contains an origin for replication of the bacterial chromosome.
D)It allows a researcher to distinguish plasmids with an insert from those without an insert.
E)It is required for packing plasmid DNA into bacteriophage heads.
A)It provides several unique restriction enzyme recognition sites for opening up the plasmid.
B)It allows a researcher to separate bacterial cells with a plasmid from those without a plasmid.
C)It contains an origin for replication of the bacterial chromosome.
D)It allows a researcher to distinguish plasmids with an insert from those without an insert.
E)It is required for packing plasmid DNA into bacteriophage heads.

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
16
The recognition site for a restriction enzyme is 5′ ACC^GGT 3′.The ^ symbol indicates the site of cleavage.After digestion of DNA with this restriction enzyme, how will the ends of the resulting DNA fragments look?
A)They will have sticky, single-stranded ends with 5′ overhangs.
B)They will have sticky, single-stranded ends with 3′ overhangs.
C)They will have blunt ends.
D)They will have sticky, double-stranded ends.
E)The result cannot be predicted using only this information.
A)They will have sticky, single-stranded ends with 5′ overhangs.
B)They will have sticky, single-stranded ends with 3′ overhangs.
C)They will have blunt ends.
D)They will have sticky, double-stranded ends.
E)The result cannot be predicted using only this information.

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
17
The recognition site for a restriction enzyme is 5′ ACCGG^T 3′.The ^ symbol indicates the site of cleavage.After digestion of DNA with this restriction enzyme, how will the ends of the resulting DNA fragments look?
A)They will have sticky, single-stranded ends with 5′ overhangs.
B)They will have sticky, single-stranded ends with 3′ overhangs.
C)They will have blunt ends.
D)They will have sticky, double-stranded ends.
E)The result cannot be predicted using only the information given.
A)They will have sticky, single-stranded ends with 5′ overhangs.
B)They will have sticky, single-stranded ends with 3′ overhangs.
C)They will have blunt ends.
D)They will have sticky, double-stranded ends.
E)The result cannot be predicted using only the information given.

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
18
Which vector would be best to use if you want to construct a genomic library with the largest fragments possible?
A)YAC
B)BAC
C)Plasmid
D)Virus
A)YAC
B)BAC
C)Plasmid
D)Virus

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
19
What is one difference between using restriction endonucleases and mechanical shearing of DNA?
A)Restriction endonucleases cut DNA at known specific sites while shearing occurs at random sites.
B)Restriction endonucleases cut DNA at random sites while shearing occurs at known specific sites.
C)DNA cut with restriction enzymes can be used for cloning and DNA broken by shearing cannot.
D)Restriction enzymes produce larger fragments on average than shearing.
A)Restriction endonucleases cut DNA at known specific sites while shearing occurs at random sites.
B)Restriction endonucleases cut DNA at random sites while shearing occurs at known specific sites.
C)DNA cut with restriction enzymes can be used for cloning and DNA broken by shearing cannot.
D)Restriction enzymes produce larger fragments on average than shearing.

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
20
What is the origin and physiological function of restriction endonucleases?
A)Bacteriophage produce them to gain resistance to antibiotics.
B)Restriction endonucleases are required for DNA replication in yeast.
C)Bacteria produce them to protect against viral invasion.
D)Scientists invented them and synthesize them in labs.
A)Bacteriophage produce them to gain resistance to antibiotics.
B)Restriction endonucleases are required for DNA replication in yeast.
C)Bacteria produce them to protect against viral invasion.
D)Scientists invented them and synthesize them in labs.

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
21
Why is it necessary to obtain overlapping sequences when sequencing a genome for the first time?
A)To assemble the whole genome sequence virtually from random fragments
B)To quantify the amount of DNA in the genome
C)To make sure all the clones in a library are used
D)To allow for sequencing errors to be corrected
A)To assemble the whole genome sequence virtually from random fragments
B)To quantify the amount of DNA in the genome
C)To make sure all the clones in a library are used
D)To allow for sequencing errors to be corrected

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
22
The sequence of gene B from another baby frog who is homozygous for allele b2 is shown.What effect does the mutation in allele b2 have on the protein? 5′ AGGTCGCATAAATGTTCCTGTAATTTGG… 3′
A)Allele b2 has a frameshift mutation-a deletion of one nucleotide resulting in a frameshift that changes all amino acids from that point on.
B)Allele b2 has a deletion of one nucleotide outside of the coding region and there is no effect on the protein.
C)Allele b2 has a missense mutation-a nucleotide substitution that changes one amino acid to another.
D)Allele b2 has a nonsense mutation-a nucleotide substitution that forms a stop codon.
E)Allele b2 has a silent mutation-a nucleotide substitution that does not change the protein.
A)Allele b2 has a frameshift mutation-a deletion of one nucleotide resulting in a frameshift that changes all amino acids from that point on.
B)Allele b2 has a deletion of one nucleotide outside of the coding region and there is no effect on the protein.
C)Allele b2 has a missense mutation-a nucleotide substitution that changes one amino acid to another.
D)Allele b2 has a nonsense mutation-a nucleotide substitution that forms a stop codon.
E)Allele b2 has a silent mutation-a nucleotide substitution that does not change the protein.

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
23
Baby frog #1 is homozygous for a mutant allele of gene B (allele b1).Compare the DNA sequence from baby frog #1 with the wild-type sequence to determine how allele b1 is different from the wild-type.What effect does the mutation in allele b1 have on the protein? 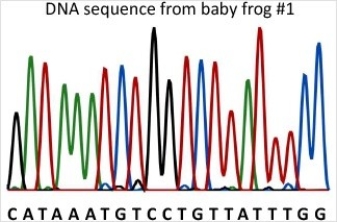
A)Allele b1 has a frameshift mutation-a deletion of one nucleotide resulting in a frameshift that changes all amino acids from that point on.
B)Allele b1 has a deletion of one nucleotide outside of the coding region and there is not effect on the protein.
C)Allele b1 has a missense mutation-a nucleotide substitution that changes one amino acid to another.
D)Allele b1 has a nonsense mutation-a nucleotide substitution that forms a stop codon.
E)Allele b1 has a nucleotide substitution that does not change the protein.

A)Allele b1 has a frameshift mutation-a deletion of one nucleotide resulting in a frameshift that changes all amino acids from that point on.
B)Allele b1 has a deletion of one nucleotide outside of the coding region and there is not effect on the protein.
C)Allele b1 has a missense mutation-a nucleotide substitution that changes one amino acid to another.
D)Allele b1 has a nonsense mutation-a nucleotide substitution that forms a stop codon.
E)Allele b1 has a nucleotide substitution that does not change the protein.

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
24
A third mutant allele of gene B (allele b3)causes the frogs' eyes to become green, instead of their normal red color.Normally, gene B effects only skin color.What term describes this allele?
A)Hypermorphic
B)Neomorphic
C)Hypomorphic
D)Amorphic
E)Haploinsufficient
A)Hypermorphic
B)Neomorphic
C)Hypomorphic
D)Amorphic
E)Haploinsufficient

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
25
Which method of fragmenting DNA would not be useful for sequencing a genome for the first time?
A)Complete digestion with a restriction enzyme
B)Partial digestion with a restriction enzyme
C)Forcing DNA through a thin needle
D)Exposing DNA to ultrasound energy
A)Complete digestion with a restriction enzyme
B)Partial digestion with a restriction enzyme
C)Forcing DNA through a thin needle
D)Exposing DNA to ultrasound energy

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
26
Why does a genomic library need more clones than what would be estimated by calculations to be complete?
A)There is a significant probability that some sequences would not be represented in a library if it was only the calculated size.
B)The restriction enzyme that is used to prepare the DNA for cloning will not digest certain sequences within the DNA.
C)The restriction enzyme used to prepare the DNA for cloning sometimes leaves a different sticky end.
D)Restriction enzymes cleave DNA preferentially at the ends of open reading frames so coding regions are preferentially cloned.
A)There is a significant probability that some sequences would not be represented in a library if it was only the calculated size.
B)The restriction enzyme that is used to prepare the DNA for cloning will not digest certain sequences within the DNA.
C)The restriction enzyme used to prepare the DNA for cloning sometimes leaves a different sticky end.
D)Restriction enzymes cleave DNA preferentially at the ends of open reading frames so coding regions are preferentially cloned.

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
27
In frogs, green skin is determined by gene B.The wild-type sequence of the 5′ end of the RNA-like strand of the entire first exon is shown below.The gene encodes an enzyme that functions to convert a brown compound into a green pigment in the skin.
5′ ACTCAAGCACAGGTCGCATAAATGTTCCTGTTATTTGG… 3′
The table of codons is provided here: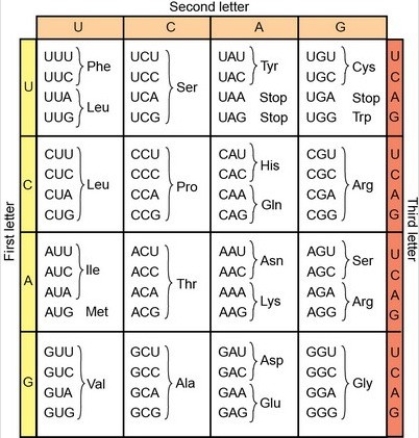
Which are the first three amino acids encoded by the wild-type sequence of gene B?
A)N-Thr-Gln-Ala…
B)N-Pro-Asn-Asn…
C)N-Met-Phe-Leu…
D)N-Ser-Ser-Val…
E)The correct sequence is not shown here.
5′ ACTCAAGCACAGGTCGCATAAATGTTCCTGTTATTTGG… 3′
The table of codons is provided here:

Which are the first three amino acids encoded by the wild-type sequence of gene B?
A)N-Thr-Gln-Ala…
B)N-Pro-Asn-Asn…
C)N-Met-Phe-Leu…
D)N-Ser-Ser-Val…
E)The correct sequence is not shown here.

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
28
Baby frog #1 was born with brown skin instead of the normal green.DNA was collected from the baby frog and the DNA sequence of part of gene B was determined using the primer 5′ ACTCAAGCACAGGTCG 3′.What is the entire sequence of the smallest DNA fragment produced in the sequencing reaction? (See diagram for sequencing data. ) https://commons.wikimedia.org/wiki/File:DNA_sequence.svg 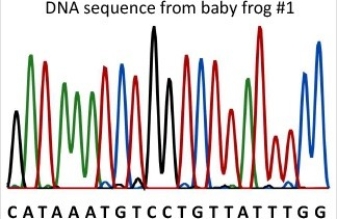
A)C
B)5′ ACTCAAGCACAGGTCGC
C)5′ AGGTCGG
D)5′ CATAAATGTCCTGTTATTTGG
E)5′ CGACCTG

A)C
B)5′ ACTCAAGCACAGGTCGC
C)5′ AGGTCGG
D)5′ CATAAATGTCCTGTTATTTGG
E)5′ CGACCTG

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck
29
Why was another nucleotide not added to the smallest fragment?
A)DNA polymerase became inactive after adding the last nucleotide.
B)The last nucleotide added does not have a 3′ hydroxyl group.
C)The last nucleotide added does not have a 5′ phosphate group.
D)The DNA strand reached a length where it cannot grow any longer.
E)DNA polymerase reached the end of the template.
A)DNA polymerase became inactive after adding the last nucleotide.
B)The last nucleotide added does not have a 3′ hydroxyl group.
C)The last nucleotide added does not have a 5′ phosphate group.
D)The DNA strand reached a length where it cannot grow any longer.
E)DNA polymerase reached the end of the template.

Unlock Deck
Unlock for access to all 29 flashcards in this deck.
Unlock Deck
k this deck


