Deck 19: Genomes and Proteomes
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Match between columns
Premises:
RNA-seq
RNA-seq
RNA-seq
Cellular proteomics
Cellular proteomics
Cellular proteomics
PCR
PCR
PCR
Responses:
mRNA/cDNA
DNA
protein
mRNA/cDNA
DNA
protein
mRNA/cDNA
DNA
protein
mRNA/cDNA
DNA
protein
mRNA/cDNA
DNA
protein
mRNA/cDNA
DNA
protein
mRNA/cDNA
DNA
protein
mRNA/cDNA
DNA
protein
mRNA/cDNA
DNA
protein
Question
Question
Match between columns
Question
Question
Question
Question
Match between columns
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/81
Play
Full screen (f)
Deck 19: Genomes and Proteomes
1
Which domain(s) have densely packed genomes with little noncoding space?
A) Archaea only
B) Bacteria only
C) Eukarya only
D) Archaea and Eukarya
E) Bacteria and Archaea
A) Archaea only
B) Bacteria only
C) Eukarya only
D) Archaea and Eukarya
E) Bacteria and Archaea
E
2
What marks the 5 ' end of an ORF in a DNA sequence?
A) the stop codon TAG
B) the stop codon TAA
C) the start codon AUG
D) the start codon ATG
E) the promoter region containing a consensus sequence
A) the stop codon TAG
B) the stop codon TAA
C) the start codon AUG
D) the start codon ATG
E) the promoter region containing a consensus sequence
D
3
The chemical difference between dideoxyribonucleotides used in Sanger sequencing and the deoxyribonucleotides normally found in DNA is the presence of a(n) _____ in the deoxyribose sugar of dideoxyribonucleotides.
A) 3' - H rather than - OH
B) 5' - H rather than - OH
C) 5' - OH rather than - H
D) 5' - H rather than - OH
E) 3' - COOH rather than - OH
A) 3' - H rather than - OH
B) 5' - H rather than - OH
C) 5' - OH rather than - H
D) 5' - H rather than - OH
E) 3' - COOH rather than - OH
A
4
In whole-genome shotgun sequencing, ____.
A) DNA is broken into many random, overlapping fragments that are sequenced and then assembled using computer algorithms
B) whole chromosomes are sequenced intact from the 3' end to the 5' end
C) whole chromosomes are sequenced intact from the 5' end to the 3' end
D) DNA is broken into a few, non-overlapping fragments that can be read directly by computer algorithms
E) DNA is broken into individual nucleotides that are sequenced and then assembled using computer algorithms
A) DNA is broken into many random, overlapping fragments that are sequenced and then assembled using computer algorithms
B) whole chromosomes are sequenced intact from the 3' end to the 5' end
C) whole chromosomes are sequenced intact from the 5' end to the 3' end
D) DNA is broken into a few, non-overlapping fragments that can be read directly by computer algorithms
E) DNA is broken into individual nucleotides that are sequenced and then assembled using computer algorithms

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
5
To study how genes have evolved, scientists compare the genome sequences of related organisms, a research approach known as _____.
A) genome sequence determination
B) genome annotation
C) comparative genomics
D) functional genomics
E) proteomics
A) genome sequence determination
B) genome annotation
C) comparative genomics
D) functional genomics
E) proteomics

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
6
How do scientists most often access genome sequences determined by research projects?
A) They must request the sequence from the group that performed the research.
B) They must request the sequence from the governmental or private group that funded the research.
C) They must request the sequence from the company that performed the sequencing.
D) They can retrieve the sequence from an online public database, such as GenBank.
E) They must sequence the organism themselves to obtain the sequence.
A) They must request the sequence from the group that performed the research.
B) They must request the sequence from the governmental or private group that funded the research.
C) They must request the sequence from the company that performed the sequencing.
D) They can retrieve the sequence from an online public database, such as GenBank.
E) They must sequence the organism themselves to obtain the sequence.

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
7
Which study indicates that some mutations are inherited from our parents?
A) study of mutations in nerve cells
B) study of mutations in somatic cells
C) study of mutations in germline cells
D) study of mutations in premature infants
E) study of mutations in in vitro babies
A) study of mutations in nerve cells
B) study of mutations in somatic cells
C) study of mutations in germline cells
D) study of mutations in premature infants
E) study of mutations in in vitro babies

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
8
In the whole-genome shotgun method, the genome is fragmented and individual fragments are sequenced. How is the order of the nucleotides determined for the original intact chromosome?
A) The fragmentation is done in a systematic way such that the physical arrangement of fragments is readily apparent.
B) The random fragmentation produces overlapping sequences that can be aligned and assembled to generate the original intact DNA molecule.
C) We supplement our information with data from a different technique to determine the final chromosome arrangement.
D) DNA hybridization assays are conducted to determine the physical arrangement of the genes on the chromosome.
E) After sequencing, the fragments are labeled and used as probes in a microarray.
A) The fragmentation is done in a systematic way such that the physical arrangement of fragments is readily apparent.
B) The random fragmentation produces overlapping sequences that can be aligned and assembled to generate the original intact DNA molecule.
C) We supplement our information with data from a different technique to determine the final chromosome arrangement.
D) DNA hybridization assays are conducted to determine the physical arrangement of the genes on the chromosome.
E) After sequencing, the fragments are labeled and used as probes in a microarray.

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
9
Which element of a dideoxy sequencing reaction allows for visualization of the DNA sequence?
A) the mixture of the four dideoxyribonucleotides, each with a different fluorescent label
B) DNA primer
C) the mixture of the four deoxyribonucleotides
D) DNA polymerase
E) DNA ligase
A) the mixture of the four dideoxyribonucleotides, each with a different fluorescent label
B) DNA primer
C) the mixture of the four deoxyribonucleotides
D) DNA polymerase
E) DNA ligase

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
10
The percentage of the human genome sequence that is protein-coding is less than _____.
A) 2%
B) 20%
C) 30%
D) 50%
E) 85%
A) 2%
B) 20%
C) 30%
D) 50%
E) 85%

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
11
The human genome is composed of approximately _____ billion base pairs.
A) 1
B) 3
C) 7
D) 10
E) 14
A) 1
B) 3
C) 7
D) 10
E) 14

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
12
The Human Genome Project completed in 2003 focused on sequencing the genome(s) of _____.
A) the mouse only
B) humans only
C) bacteria only
D) humans and prokaryotes only
E) humans and other model organisms
A) the mouse only
B) humans only
C) bacteria only
D) humans and prokaryotes only
E) humans and other model organisms

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
13
During a DNA sequencing experiment, the researcher adds all the required components to the reaction, but accidentally adds only one of the four dideoxynucleotides, the ddC. What products will be observed after this reaction is complete?
A) All fragments will end in A.
B) All fragments will end in C.
C) All fragments will end in G.
D) All fragments will end in T.
E) No fragments will be made.
A) All fragments will end in A.
B) All fragments will end in C.
C) All fragments will end in G.
D) All fragments will end in T.
E) No fragments will be made.

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
14
What is a difference between human male and female genomes?
A) Males have 46 chromosomes; females have 45.
B) Males have 22 pairs of autosomal chromosomes; females have 21 pairs of autosomal chromosomes.
C) Males have 23 pairs of chromosomes plus 1 Y chromosome; females have 23 pairs of chromosomes.
D) Males have 24 different chromosomes; females have 23 different chromosomes.
E) Males have an X chromosome; females do not.
A) Males have 46 chromosomes; females have 45.
B) Males have 22 pairs of autosomal chromosomes; females have 21 pairs of autosomal chromosomes.
C) Males have 23 pairs of chromosomes plus 1 Y chromosome; females have 23 pairs of chromosomes.
D) Males have 24 different chromosomes; females have 23 different chromosomes.
E) Males have an X chromosome; females do not.

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
15
A scientist obtains the DNA sequence data for a bacterial plasmid. The data include As, Ts, and Gs, but no Cs. What is the most likely cause of this result?
A) ddCs were omitted from the sequencing reaction.
B) The cytosine deoxyribonucleotides were omitted from the sequencing reaction.
C) DNA polymerase was omitted from the sequencing reaction mix.
D) The ddG fluorescent tags were degraded.
E) There is no cytosine in this DNA molecule.
A) ddCs were omitted from the sequencing reaction.
B) The cytosine deoxyribonucleotides were omitted from the sequencing reaction.
C) DNA polymerase was omitted from the sequencing reaction mix.
D) The ddG fluorescent tags were degraded.
E) There is no cytosine in this DNA molecule.

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
16
Deoxyribonucleotides are included in the Sanger sequencing reaction mix to _____.
A) provide a substrate for RNA polymerase
B) enhance the activity of the dideoxyribonucleotides
C) allow longer DNA fragments (up to 300 additional base pairs) to be sequenced
D) allow the production of multiple DNA fragments of different lengths
E) prevent the DNA polymerase from making too many errors
A) provide a substrate for RNA polymerase
B) enhance the activity of the dideoxyribonucleotides
C) allow longer DNA fragments (up to 300 additional base pairs) to be sequenced
D) allow the production of multiple DNA fragments of different lengths
E) prevent the DNA polymerase from making too many errors

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
17
When the human genome was sequenced, we learned that there are fewer than expected protein-coding genes (approximately 20,000). Yet, the total number of proteins produced in humans approaches 100,000. What accounts for this discrepancy in numbers?
A) We have not yet identified all of the open reading frames in the human genome.
B) We have not yet fully sequenced the human genome.
C) Processing mechanisms for mRNAs allow multiple proteins to be produced from a single DNA sequence.
D) Some noncoding DNA sequences encode proteins.
E) There has been a gross over-estimation of the number of proteins produced in humans.
A) We have not yet identified all of the open reading frames in the human genome.
B) We have not yet fully sequenced the human genome.
C) Processing mechanisms for mRNAs allow multiple proteins to be produced from a single DNA sequence.
D) Some noncoding DNA sequences encode proteins.
E) There has been a gross over-estimation of the number of proteins produced in humans.

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
18
What is an open reading frame (ORF)?
A) a protein coding sequence plus associated regulatory sequences
B) the sequence between and including a start codon and a stop codon
C) the sequence between and including a start codon and a stop codon, minus the introns
D) the sequence between and including a start codon and a stop codon, minus the exons
E) the sequence between a start codon and stop codon in prokaryotes only
A) a protein coding sequence plus associated regulatory sequences
B) the sequence between and including a start codon and a stop codon
C) the sequence between and including a start codon and a stop codon, minus the introns
D) the sequence between and including a start codon and a stop codon, minus the exons
E) the sequence between a start codon and stop codon in prokaryotes only

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
19
In humans, the majority of a transcription unit is composed of ____.
A) exons
B) introns
C) promoter regions
D) start codons
E) regulatory elements
A) exons
B) introns
C) promoter regions
D) start codons
E) regulatory elements

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
20
Which pre-mRNA processing mechanism allows different proteins to be produced from the same DNA sequence?
A) transposition
B) alternative splicing
C) intron shuffling
D) intron splicing
E) open reading frame shuffling
A) transposition
B) alternative splicing
C) intron shuffling
D) intron splicing
E) open reading frame shuffling

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
21
In the microarray shown in the textbook (p. 414), the cDNAs were labeled with red and green fluorescent tags. Why do some spots on the microarray emit yellow light?
A) Red and green are two of the primary colors, and if both cDNAs hybridize to the spot, the combination of the two will produce yellow light.
B) The yellow light comes from the laser, and a yellow spot indicates that neither cDNA hybridized to that spot on the microarray.
C) The over-expression of one cDNA relative to the other will skew the color pattern of the spot on the microarray, resulting in the yellow color.
D) The color choice was an arbitrary decision by the artist and doesn't reflect how the process actually works.
E) Yellow spots are those that contain no DNA probes on the chip.
A) Red and green are two of the primary colors, and if both cDNAs hybridize to the spot, the combination of the two will produce yellow light.
B) The yellow light comes from the laser, and a yellow spot indicates that neither cDNA hybridized to that spot on the microarray.
C) The over-expression of one cDNA relative to the other will skew the color pattern of the spot on the microarray, resulting in the yellow color.
D) The color choice was an arbitrary decision by the artist and doesn't reflect how the process actually works.
E) Yellow spots are those that contain no DNA probes on the chip.

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
22
Which technique can most accurately quantify RNA and determine levels of gene expression?
A) RNA-seq
B) microarray analysis
C) Illumina/Solexa sequencing
D) alignment searches
E) Sanger sequencing
A) RNA-seq
B) microarray analysis
C) Illumina/Solexa sequencing
D) alignment searches
E) Sanger sequencing

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
23
What can be removed from pre-mRNA during alternative splicing?
A) exons and ribosomal binding sites
B) ribosomal binding sites
C) exons only
D) introns only
E) exons and introns
A) exons and ribosomal binding sites
B) ribosomal binding sites
C) exons only
D) introns only
E) exons and introns

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
24
RNAi is commonly used in which experimental strategy?
A) gene overexpression
B) DNA sequencing
C) gene knockout
D) gene knockdown
E) microarray analysis
A) gene overexpression
B) DNA sequencing
C) gene knockout
D) gene knockdown
E) microarray analysis

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
25
A researcher wants to investigate the function of a gene without permanently altering the gene's function or activity. Which experimental approach should she choose?
A) gene knockout
B) gene knockdown
C) electrophoresis
D) RNA-seq
E) bioinformatics
A) gene knockout
B) gene knockdown
C) electrophoresis
D) RNA-seq
E) bioinformatics

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
26
During DNA sequence annotation, which feature is not identified?
A) protein-coding genes
B) noncoding RNA genes
C) origins of replication
D) pseudogenes
E) protein interactions
A) protein-coding genes
B) noncoding RNA genes
C) origins of replication
D) pseudogenes
E) protein interactions

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
27
Microarrays let us identify which DNA sequences are present in a particular cell type under certain conditions. How can microarrays help us understand cellular functions?
A) Microarrays let us study different cellular structures under different conditions.
B) Microarrays let us directly measure protein expression in individual cells.
C) Microarrays let us identify which portions of a genome were being expressed in a cell at a particular time.
D) Microarrays let us identify which DNA sequences are present in a particular cell type under certain conditions.
E) Microarrays let us identify which portions of a genome serve as regulatory sequences.
A) Microarrays let us study different cellular structures under different conditions.
B) Microarrays let us directly measure protein expression in individual cells.
C) Microarrays let us identify which portions of a genome were being expressed in a cell at a particular time.
D) Microarrays let us identify which DNA sequences are present in a particular cell type under certain conditions.
E) Microarrays let us identify which portions of a genome serve as regulatory sequences.

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
28
The study of gene function by observing changes in phenotype is termed _____.
A) phenomics
B) genomics
C) mutagenomics
D) proteomics
E) transcriptomics
A) phenomics
B) genomics
C) mutagenomics
D) proteomics
E) transcriptomics

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
29
Through a sequence similarity search, a researcher identifies a gene whose predicted protein product has a putative function of a photosynthesis-enhancing enzyme. Which experimental method could be performed to verify this function?
A) PCR
B) an assay measuring photosynthetic productivity
C) identification of signature genes
D) DNA microarray
E) gene knockout
A) PCR
B) an assay measuring photosynthetic productivity
C) identification of signature genes
D) DNA microarray
E) gene knockout

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
30
Which eukaryotic organelle(s) contain its/their own DNA?
A) chloroplasts only
B) endoplasmic reticulum only
C) mitochondria only
D) mitochondria and chloroplasts
E) mitochondria and endoplasmic reticulum
A) chloroplasts only
B) endoplasmic reticulum only
C) mitochondria only
D) mitochondria and chloroplasts
E) mitochondria and endoplasmic reticulum

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
31
Which field of study applies mathematics and computer science to gain information from biological data, such as data related to gene structure and function?
A) computer forensics
B) statistical biology
C) bioinformatics
D) genome sampling
E) metagenomics
A) computer forensics
B) statistical biology
C) bioinformatics
D) genome sampling
E) metagenomics

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
32
Many cancers can be identified based on microarray analysis of _____.
A) signature genes
B) genes in red blood cells
C) mitochondrial DNA
D) Y chromosome DNA
E) DNA probes
A) signature genes
B) genes in red blood cells
C) mitochondrial DNA
D) Y chromosome DNA
E) DNA probes

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
33
A(n) _____ has a DNA sequence similar to a functional gene, but has been mutated so that it no longer produces a functional gene product.
A) pseudogene
B) homologous gene
C) orthologous gene
D) microRNA-encoding sequence
E) operon
A) pseudogene
B) homologous gene
C) orthologous gene
D) microRNA-encoding sequence
E) operon

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
34
Protein-coding ORFs are easier to identify in which type of organism?
A) eukaryotes because they have easily identifiable regulatory regions
B) eukaryotes because they have fewer introns
C) prokaryotes because they have few introns
D) eukaryotes because they have more exons
E) prokaryotes because they have more exons
A) eukaryotes because they have easily identifiable regulatory regions
B) eukaryotes because they have fewer introns
C) prokaryotes because they have few introns
D) eukaryotes because they have more exons
E) prokaryotes because they have more exons

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
35
RNA-seq (whole-transcriptome sequencing) _____.
A) is a new hybridization technique used in transcriptomics
B) is the most common method used to identify certain cancer genes
C) identifies and quantifies RNA transcripts in a sample
D) is being replaced by DNA microarrays in transcriptomics
E) can only identify 100 sequences at a time
A) is a new hybridization technique used in transcriptomics
B) is the most common method used to identify certain cancer genes
C) identifies and quantifies RNA transcripts in a sample
D) is being replaced by DNA microarrays in transcriptomics
E) can only identify 100 sequences at a time

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
36
What is directly sequenced in RNA-seq?
A) protein
B) transcripts
C) RNA
D) DNA
E) cDNA
A) protein
B) transcripts
C) RNA
D) DNA
E) cDNA

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
37
A gene encoding a subunit of hemoglobin in humans is found to be homologous to a gene in chimpanzees. The homologous gene in chimpanzees likely _____.
A) is a pseudogene
B) encodes the same hemoglobin subunit
C) is inactive because it is in a different species
D) is a regulatory sequence
E) encodes a respiratory enzyme
A) is a pseudogene
B) encodes the same hemoglobin subunit
C) is inactive because it is in a different species
D) is a regulatory sequence
E) encodes a respiratory enzyme

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
38
A sequence similarity search reveals that a candidate gene's sequence has a partial match to a known gene's sequence. What does this most likely indicate?
A) There is no relationship between the candidate and known gene.
B) The candidate gene encodes an identical transcript to the known gene.
C) The candidate gene encodes the same protein as the known gene.
D) The protein encoded by the candidate gene contains a protein domain also found in the known gene's protein.
E) The protein encoded by the candidate gene has the same function as the protein encoded by the known gene.
A) There is no relationship between the candidate and known gene.
B) The candidate gene encodes an identical transcript to the known gene.
C) The candidate gene encodes the same protein as the known gene.
D) The protein encoded by the candidate gene contains a protein domain also found in the known gene's protein.
E) The protein encoded by the candidate gene has the same function as the protein encoded by the known gene.

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
39
Which organism has the largest number of protein-coding genes?
A) E. coli (bacteria)
B) Saccharomyces cerevisiae (yeast)
C) Homo sapiens (human)
D) Oryza sativa (rice)
E) Drosophila melanogaster (fruit fly)
A) E. coli (bacteria)
B) Saccharomyces cerevisiae (yeast)
C) Homo sapiens (human)
D) Oryza sativa (rice)
E) Drosophila melanogaster (fruit fly)

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
40
A researcher obtains the results of his RNA-seq analysis. Several transcripts are listed as being derived from the same gene. How is this possible?
A) Alternative splicing allows for different transcripts to be created by the same gene sequence.
B) The researcher's sample was contaminated.
C) Exon shuffling allows for different transcripts to be created by the same gene sequence.
D) Intron shuffling allows for different transcripts to be created by the same gene sequence.
E) It is not possible; there was an error in the alignment process.
A) Alternative splicing allows for different transcripts to be created by the same gene sequence.
B) The researcher's sample was contaminated.
C) Exon shuffling allows for different transcripts to be created by the same gene sequence.
D) Intron shuffling allows for different transcripts to be created by the same gene sequence.
E) It is not possible; there was an error in the alignment process.

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
41
The _____ technique is most effective for transcriptomics studies in non-model organisms because _____.
A) DNA microarray; DNA sequences from related organisms can be used as probes
B) RNA-seq; it is less expensive
C) DNA microarray; prior knowledge about the genome sequence is not required
D) RNA-seq; the probes used are smaller than those for DNA microarrays
E) DNA microarray; fluorescence gives the best indication of gene activity
A) DNA microarray; DNA sequences from related organisms can be used as probes
B) RNA-seq; it is less expensive
C) DNA microarray; prior knowledge about the genome sequence is not required
D) RNA-seq; the probes used are smaller than those for DNA microarrays
E) DNA microarray; fluorescence gives the best indication of gene activity

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
42
Gene duplication can result from _____.
A) a normal crossover event
B) exon shuffling
C) unequal crossing-over
D) a point mutation
E) alternative splicing
A) a normal crossover event
B) exon shuffling
C) unequal crossing-over
D) a point mutation
E) alternative splicing

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
43
How might a researcher best determine the location of a protein in a cell?
A) studying the amino acid sequence of the protein
B) attaching a fluorescent tag to the protein in some way to visualize its cellular location under a microscope
C) attaching an antibiotic tag to the protein and determining its location using DNA microarrays
D) analyzing the untranslated regions of the gene encoding the protein
E) understanding the protein's function
A) studying the amino acid sequence of the protein
B) attaching a fluorescent tag to the protein in some way to visualize its cellular location under a microscope
C) attaching an antibiotic tag to the protein and determining its location using DNA microarrays
D) analyzing the untranslated regions of the gene encoding the protein
E) understanding the protein's function

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
44
DNA microarrays that reveal differential expression patterns are used to study the _____.
A) genome
B) transcriptome
C) proteome
D) interactome
E) ribosome
A) genome
B) transcriptome
C) proteome
D) interactome
E) ribosome

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
45
Which event can introduce a protein domain into a pre-existing protein?
A) gene alignment
B) intron shuffling
C) exon shuffling
D) peptide shuffling
E) exon splicing
A) gene alignment
B) intron shuffling
C) exon shuffling
D) peptide shuffling
E) exon splicing

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
46
To experimentally determine the function of an annotated gene, a researcher may _____.
A) look for similar gene sequences of known function
B) examine the putative protein structure
C) perform a gene knockout
D) find a pseudogene for the gene
E) determine the amino acid sequence
A) look for similar gene sequences of known function
B) examine the putative protein structure
C) perform a gene knockout
D) find a pseudogene for the gene
E) determine the amino acid sequence

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
47
Unequal crossing over _____.
A) is an abnormal crossover event that occurs during mitosis
B) occurs between different points on homologous chromosomes
C) generates a single point mutation
D) is the result of alternative splicing
E) occurs between heterologous chromosomes
A) is an abnormal crossover event that occurs during mitosis
B) occurs between different points on homologous chromosomes
C) generates a single point mutation
D) is the result of alternative splicing
E) occurs between heterologous chromosomes

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
48
When copies of a gene are found at different locations on the same or different chromosomes, the phenomenon is called _____.
A) dispersed duplication
B) dispersed localization
C) duplicate localization
D) gene dispersion
E) random dispersion
A) dispersed duplication
B) dispersed localization
C) duplicate localization
D) gene dispersion
E) random dispersion

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
49
The proteome represents _____, while the cellular proteome represents _____.
A) all of the proteins able to be made by all living organisms; the proteins made by unicellular organisms
B) the set of proteins made by any multicellular organism; the set of proteins made by unicellular organisms
C) all of the proteins able to be expressed by an organism's genome; the subset of proteins found in a particular cell type
D) all of the proteins that are common to all living organisms; the proteins found in the same cell types of different species
E) the subject of proteomics research; the subject of phenomics research
A) all of the proteins able to be made by all living organisms; the proteins made by unicellular organisms
B) the set of proteins made by any multicellular organism; the set of proteins made by unicellular organisms
C) all of the proteins able to be expressed by an organism's genome; the subset of proteins found in a particular cell type
D) all of the proteins that are common to all living organisms; the proteins found in the same cell types of different species
E) the subject of proteomics research; the subject of phenomics research

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
50
Researcher A studies the protein interactions involving the polypeptides that comprise hemoglobin, while Researcher B studies protein interactions between a protein kinase and its target protein. Researcher A will therefore focus on ____, and researcher B will focus on ____.
A) protein localization; protein sequence
B) protein sequence; protein localization
C) permanent protein interactions; transient protein interactions
D) transient protein interactions; permanent protein interactions
E) protein regulation; transcriptional regulation
A) protein localization; protein sequence
B) protein sequence; protein localization
C) permanent protein interactions; transient protein interactions
D) transient protein interactions; permanent protein interactions
E) protein regulation; transcriptional regulation

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
51
In humans, there are three genes encoding different nitric oxide synthase enzymes. These three genes form a ____.
A) multigene locus
B) replicate gene family
C) multi-enzyme family
D) multigene family
E) gene grouping
A) multigene locus
B) replicate gene family
C) multi-enzyme family
D) multigene family
E) gene grouping

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
52
Proteomics _____.
A) refers to the analysis of the entire protein content of a cell
B) refers to the analysis of all the DNA of a species
C) looks only at plasmids
D) studies mRNA levels
E) uses DNA chips
A) refers to the analysis of the entire protein content of a cell
B) refers to the analysis of all the DNA of a species
C) looks only at plasmids
D) studies mRNA levels
E) uses DNA chips

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
53
During a microarray analysis, cDNAs made from normal cells are stained with a green fluor and cDNAs from abnormal cells are stained with a red fluor. Which of the following statements is true about the results of this experiment?
A) Red spots indicate genes that are under-expressed in abnormal cells.
B) Red spots indicate genes that are not expressed in abnormal cells.
C) Red spots indicate genes that are over-expressed in abnormal cells.
D) Red spots indicate genes that are over-expressed in normal cells.
E) Red spots indicate pseudogenes.
A) Red spots indicate genes that are under-expressed in abnormal cells.
B) Red spots indicate genes that are not expressed in abnormal cells.
C) Red spots indicate genes that are over-expressed in abnormal cells.
D) Red spots indicate genes that are over-expressed in normal cells.
E) Red spots indicate pseudogenes.

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
54
What has been learned from comparative genomics studies?
A) Similar genes in different organisms perform very different functions.
B) Prokaryotic DNA has different nucleotide bases compared to eukaryotic DNA.
C) Similar genes do not exist in different species.
D) Some genes are present in the genomes of almost all present-day organisms.
E) Different species can have identical genome sequences.
A) Similar genes in different organisms perform very different functions.
B) Prokaryotic DNA has different nucleotide bases compared to eukaryotic DNA.
C) Similar genes do not exist in different species.
D) Some genes are present in the genomes of almost all present-day organisms.
E) Different species can have identical genome sequences.

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
55
Which gene would most likely be found in the genomes of all present-day organisms?
A) an amino-acyl tRNA synthetase gene
B) a photosynthetic gene
C) a nitrogen fixation gene
D) a gene for anaerobic respiration-producing lactate
E) a gene for cellulose breakdown
A) an amino-acyl tRNA synthetase gene
B) a photosynthetic gene
C) a nitrogen fixation gene
D) a gene for anaerobic respiration-producing lactate
E) a gene for cellulose breakdown

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
56
Multigene families arise from _____.
A) a single gene duplication event
B) repeated cycles of gene duplication followed by mutation
C) single point mutations
D) exon shuffling
E) deletions of genes
A) a single gene duplication event
B) repeated cycles of gene duplication followed by mutation
C) single point mutations
D) exon shuffling
E) deletions of genes

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
57
The goal(s) of proteomics include(s) the determination of protein_____.
A) structure only
B) function only
C) location only
D) sequence, interactions, and structure
E) structure, function, location, and interactions
A) structure only
B) function only
C) location only
D) sequence, interactions, and structure
E) structure, function, location, and interactions

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
58
Protein structure can be predicted nonexperimentally by ____, but not as reliably as by experimental methods.
A) microscopy
B) nuclear magnetic resonance
C) X-ray crystallography
D) computer algorithms based on amino acid chemical attractions
E) electrophoresis
A) microscopy
B) nuclear magnetic resonance
C) X-ray crystallography
D) computer algorithms based on amino acid chemical attractions
E) electrophoresis

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
59
Genes that are closely related evolutionarily, have the same function, and are present in the genomes of two or more different organisms are called _____.
A) genologs
B) heterologs
C) Paralogs
D) Metalogs
E) orthologs
A) genologs
B) heterologs
C) Paralogs
D) Metalogs
E) orthologs

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
60
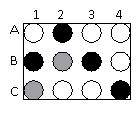
A portion of a microarray is shown above in which the expression of genes on human chromosome 21 was examined in normal brain cells (cDNAs labeled with a white fluor) or brain cells from an individual with a neurological disorder (cDNAs labeled with a black fluor).
Which genes are over-expressed in the abnormal cells compared to the normal cells?
A) A2, B1, B3, and C4
B) B2 and C1
C) A1, A3, A4, B4, C2, and C3
D) A1, A2, B2, and C1
E) A2, C2, and C4

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
61
Match between columns
Premises:
RNA-seq
RNA-seq
RNA-seq
Cellular proteomics
Cellular proteomics
Cellular proteomics
PCR
PCR
PCR
Responses:
mRNA/cDNA
DNA
protein
mRNA/cDNA
DNA
protein
mRNA/cDNA
DNA
protein
mRNA/cDNA
DNA
protein
mRNA/cDNA
DNA
protein
mRNA/cDNA
DNA
protein
mRNA/cDNA
DNA
protein
mRNA/cDNA
DNA
protein
mRNA/cDNA
DNA
protein

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
62
You are studying a particular gene in mice. In one sample of the genome from a mouse, you find that the gene that you have been studying has moved to a completely different location in the genome. The gene that has moved to the new location is bounded by inverted repeat sequences. What is responsible for this finding?
A) unequal crossing-over
B) a retrotransposon
C) a chromosomal duplication
D) a chromosomal inversion
E) a DNA transposon
A) unequal crossing-over
B) a retrotransposon
C) a chromosomal duplication
D) a chromosomal inversion
E) a DNA transposon

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
63
Match between columns

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
64
Human and chimpanzee genomes share 96% DNA sequence identity. What most likely causes humans to be phenotypically distinct from chimpanzees?
A) the presence of many new genes unique to humans
B) a much larger human genome compared to the chimpanzee genome
C) the loss of higher functioning genes in chimpanzees
D) dramatic changes in chromosome structure and organization in humans
E) subtle mutations in protein coding sequences and changes in regulatory units of human genes
A) the presence of many new genes unique to humans
B) a much larger human genome compared to the chimpanzee genome
C) the loss of higher functioning genes in chimpanzees
D) dramatic changes in chromosome structure and organization in humans
E) subtle mutations in protein coding sequences and changes in regulatory units of human genes

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
65
Genes with novel functions are most commonly produced by ____.
A) inversion of DNA sequences
B) nondisjunction during meiosis
C) unequal crossing-over
D) exon shuffling
E) small mutations
A) inversion of DNA sequences
B) nondisjunction during meiosis
C) unequal crossing-over
D) exon shuffling
E) small mutations

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
66
Which of the following is NOT a typical characteristic of a prokaryotic genome like that of E. coli ?
A) The genes are close together with little space between them.
B) The genes almost always contain introns.
C) Genes can be transcribed off either of the two strands of DNA.
D) Some genes are single transcription units, but about 50% of the protein-coding genes are found in operons.
E) The genes vary in length and reflect the lengths of their encoded proteins.
A) The genes are close together with little space between them.
B) The genes almost always contain introns.
C) Genes can be transcribed off either of the two strands of DNA.
D) Some genes are single transcription units, but about 50% of the protein-coding genes are found in operons.
E) The genes vary in length and reflect the lengths of their encoded proteins.

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
67
Match between columns

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
68
Summarize the relationships between the following types of organisms with respect to genome size: viruses, bacteria, archaea, and eukaryotes.

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
69
Which gene-altering event is most likely to create a new gene encoding a functional protein?
A) gene duplication followed by small mutations
B) gene duplication followed by large mutations
C) exon shuffling
D) transposition of DNA sequences
E) inversion of DNA sequences
A) gene duplication followed by small mutations
B) gene duplication followed by large mutations
C) exon shuffling
D) transposition of DNA sequences
E) inversion of DNA sequences

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
70
What are the main differences between the fields of genomics, transcriptomics, and proteomics?

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
71
A researcher obtains a sequence of a gene of interest, including the ORF, 5' untranslated region, and 3 ' untranslated region. Which portion of the gene sequence encodes the protein?
A) the 5 ' untranslated region and the ORF only
B) the ORF only
C) the ORF and 3 ' untranslated region only
D) the 5 ' and 3 ' untranslated regions only
E) the ORF, 5 ' untranslated region, and 3 ' untranslated region
A) the 5 ' untranslated region and the ORF only
B) the ORF only
C) the ORF and 3 ' untranslated region only
D) the 5 ' and 3 ' untranslated regions only
E) the ORF, 5 ' untranslated region, and 3 ' untranslated region

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
72
Comparative genomics is an approach used to learn how genes and genomes have evolved. Describe three different concepts that have been revealed by comparative genomics studies.

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
73
What is the difference between a gene knockout and a gene knockdown?

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
74
Chromosomal rearrangement can occur by ____.
A) translocation only
B) inversion only
C) translocation and inversion
D) point mutations
E) alternative splicing
A) translocation only
B) inversion only
C) translocation and inversion
D) point mutations
E) alternative splicing

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
75
List at least three differences in how the E. coli and human genomes are arranged.

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
76
You are studying a particular gene in mice. In one sample of the genome from a mouse, you find two copies of the gene while in other mice you have studied there are two copies of the gene. One of the copies is in the normal location; the other is in a completely different location in the genome. The copy of the gene found in a different location from what is normal is bounded by sequences that are directly repeated rather than being in an inverted form. What is responsible for this finding?
A) a retrotransposon
B) a DNA transposon
C) a point mutation
D) a chromosomal inversion
E) an unequal cross-over
A) a retrotransposon
B) a DNA transposon
C) a point mutation
D) a chromosomal inversion
E) an unequal cross-over

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
77
There are three types of sequencing techniques described in the text: Sanger (dideoxy) sequencing, whole-genome shotgun sequencing, and Illumina/Solexa sequencing. Which one(s) would be best to use for sequencing a single gene versus an entire genome and why?

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
78
Which was the first mammalian genome sequenced?
A) chimpanzee
B) gorilla
C) mouse
D) human
E) sheep
A) chimpanzee
B) gorilla
C) mouse
D) human
E) sheep

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
79
A researcher is interested in creating a phylogenetic tree showing the evolutionary relationship of several cyanobacterial species based on the closeness of their nitrogenase gene sequences. Which field of research most directly applies to the researcher's project?
A) comparative proteomics
B) comparative genomics
C) comparative transcriptomics
D) comparative phylogenetics
E) comparative phenotype
A) comparative proteomics
B) comparative genomics
C) comparative transcriptomics
D) comparative phylogenetics
E) comparative phenotype

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck
80
Explain how proteomics is considered by some to be even more important than genome sequencing.

Unlock Deck
Unlock for access to all 81 flashcards in this deck.
Unlock Deck
k this deck


