Deck 20: Genomics
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/41
Play
Full screen (f)
Deck 20: Genomics
1
Imagine that your goal is to isolate mouse mutants that stabilize microsatellite repeat sequences (i.e.,lower the frequency at which their repeat number changes).Which of the following mutations would you predict to have this stabilizing effect?
A)a mutation that reduces the rate of transcription
B)a mutation that reduces the rate of translation
C)a mutation that increases the rate of DNA synthesis
D)a mutation that reduces the occurrence of homologous recombination
E)a mutation that inactivates reverse transcriptase and/or integrase
A)a mutation that reduces the rate of transcription
B)a mutation that reduces the rate of translation
C)a mutation that increases the rate of DNA synthesis
D)a mutation that reduces the occurrence of homologous recombination
E)a mutation that inactivates reverse transcriptase and/or integrase
D
2
If the sequence of a cDNA matches a DNA sequence in the genome,then this genomic DNA is likely to
A)code for a protein.
B)code for a tRNA.
C)code for an rRNA.
D)be part of an intron.
E)be a regulatory sequence.
A)code for a protein.
B)code for a tRNA.
C)code for an rRNA.
D)be part of an intron.
E)be a regulatory sequence.
A
3
Forensic DNA fingerprinting often involves recovery of minute quantities of DNA from a crime scene.What is a concern in analyzing this DNA?
A)There is seldom enough DNA to allow PCR amplification.
B)Even with these tiny amounts of DNA,there may be too much DNA for efficient PCR.
C)Suspects may not agree to provide a sample of their DNA.
D)It is critical to avoid the introduction of contaminating human DNA unrelated to the crime.
E)Restriction enzymes may not work efficiently on the PCR-amplified DNA.
A)There is seldom enough DNA to allow PCR amplification.
B)Even with these tiny amounts of DNA,there may be too much DNA for efficient PCR.
C)Suspects may not agree to provide a sample of their DNA.
D)It is critical to avoid the introduction of contaminating human DNA unrelated to the crime.
E)Restriction enzymes may not work efficiently on the PCR-amplified DNA.
D
4
The bulk of the sequence data in whole genome sequencing comes from
A)relatively large (~ 160 kb)sequences cloned into BACs.
B)relatively small (~ 1000 base-pair)sequences cloned into plasmids.
C)whole chromosomes obtained without cloning.
D)whole chromosomes cloned into plasmids.
A)relatively large (~ 160 kb)sequences cloned into BACs.
B)relatively small (~ 1000 base-pair)sequences cloned into plasmids.
C)whole chromosomes obtained without cloning.
D)whole chromosomes cloned into plasmids.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
5
When comparing the genome sizes of a bacterial parasite versus a free-living,nonparasitic bacterium,one expects to find that
A)the proportion of G-C base pairs will be significantly higher in the parasite.
B)the proportion of G-C base pairs will be significantly lower in the parasite.
C)there will be a higher proportion of genes of unknown function in the parasite.
D)the parasite will have acquired a smaller proportion of its genes through lateral gene transfer.
E)the parasite will have a smaller genome.
A)the proportion of G-C base pairs will be significantly higher in the parasite.
B)the proportion of G-C base pairs will be significantly lower in the parasite.
C)there will be a higher proportion of genes of unknown function in the parasite.
D)the parasite will have acquired a smaller proportion of its genes through lateral gene transfer.
E)the parasite will have a smaller genome.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
6
The discipline that manages,analyzes,and interprets the vast amounts of sequence data generated from whole genome sequencing is
A)proteomics.
B)functional genomics.
C)evolutionary genomics.
D)bioinformatics.
A)proteomics.
B)functional genomics.
C)evolutionary genomics.
D)bioinformatics.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
7
One surprising discovery from the analysis of bacterial genomes is that
A)very few species contain plasmids.
B)there is only one copy of each gene present within a bacterial genome.
C)there are many genes of unknown function.
D)bacterial genomes are always contained on a single circular chromosome.
E)there is almost no difference between the genomes of diverse bacteria.
A)very few species contain plasmids.
B)there is only one copy of each gene present within a bacterial genome.
C)there are many genes of unknown function.
D)bacterial genomes are always contained on a single circular chromosome.
E)there is almost no difference between the genomes of diverse bacteria.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
8
The mechanism for the spread of LINES involves some unusual features,including
A)translation in the cytoplasm of an mRNA that was synthesized in the nucleus.
B)the nuclear localization of RNA polymerase.
C)the transport of mRNA from the cytoplasm to the nucleus.
D)the use of an enzyme that cuts DNA.
E)the use of RNA polymerase to transcribe an mRNA.
A)translation in the cytoplasm of an mRNA that was synthesized in the nucleus.
B)the nuclear localization of RNA polymerase.
C)the transport of mRNA from the cytoplasm to the nucleus.
D)the use of an enzyme that cuts DNA.
E)the use of RNA polymerase to transcribe an mRNA.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
9
In establishing paternity by DNA fingerprinting,what proportion of microsatellite repeat alleles in a child come from the father?
A)none
B)1/4
C)1/2
D)3/4
E)all
A)none
B)1/4
C)1/2
D)3/4
E)all

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
10
Which of the following makes it difficult to identify eukaryotic genes?
A)the presence of many introns
B)the large amount of DNA that does not code for proteins
C)the lack of homology between eukaryotic genes
D)A and B only
E)all of the above
A)the presence of many introns
B)the large amount of DNA that does not code for proteins
C)the lack of homology between eukaryotic genes
D)A and B only
E)all of the above

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
11
Imagine that you've sequenced the genome of a human pathogenic bacterium.In the early stages of analysis,you discover a stretch of DNA that has a significantly different GC content (the proportion of bases that are G and
A)convergent evolution.
B)that the DNA sequence analysis is in error.
C)a high rate of mutation.
D)a low rate of mutation.
E)lateral gene transfer.
A)convergent evolution.
B)that the DNA sequence analysis is in error.
C)a high rate of mutation.
D)a low rate of mutation.
E)lateral gene transfer.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
12
LINES transposable elements are closely related to
A)plasmids.
B)retroviruses.
C)poliovirus.
D)restriction enzymes.
E)DNA polymerase.
A)plasmids.
B)retroviruses.
C)poliovirus.
D)restriction enzymes.
E)DNA polymerase.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
13
In what sense are studies by 19th-century naturalists and those by early 21st-century genomic biologists similar?
A)Both focus narrowly on the underlying mechanisms of biology.
B)Both use evolutionary theory to guide their work.
C)Both take a reductionist approach by studying only one small part of a complex system.
D)Both focus on observing and describing what exists in their realms of investigation.
E)Both constantly strive to create theoretical frameworks in which to understand their findings.
A)Both focus narrowly on the underlying mechanisms of biology.
B)Both use evolutionary theory to guide their work.
C)Both take a reductionist approach by studying only one small part of a complex system.
D)Both focus on observing and describing what exists in their realms of investigation.
E)Both constantly strive to create theoretical frameworks in which to understand their findings.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
14
Sequencing eukaryotic genomes is more difficult than sequencing genomes of bacteria or archaea because of
A)the large size of eukaryotic genomes.
B)the large amount of eukaryotic repetitive DNA.
C)the high proportion of G-C base pairs in eukaryotic DNA.
D)A and B only.
E)all of the above.
A)the large size of eukaryotic genomes.
B)the large amount of eukaryotic repetitive DNA.
C)the high proportion of G-C base pairs in eukaryotic DNA.
D)A and B only.
E)all of the above.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
15
An early step in shotgun sequencing is to
A)break genomic DNA at random sites.
B)map the position of cloned DNA fragments.
C)randomly select DNA primers and hybridize these to random positions of chromosomes in preparation for sequencing.
A)break genomic DNA at random sites.
B)map the position of cloned DNA fragments.
C)randomly select DNA primers and hybridize these to random positions of chromosomes in preparation for sequencing.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
16
Homologous DNA sequences are similar sequences in two organisms that
A)are related by descent from a common ancestor.
B)are related because of convergent evolution.
C)are related by chance mutations.
D)code for identical proteins.
E)code for identical RNAs.
A)are related by descent from a common ancestor.
B)are related because of convergent evolution.
C)are related by chance mutations.
D)code for identical proteins.
E)code for identical RNAs.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
17
The goal of annotating a whole genome sequence is to
A)establish the nucleotide sequence.
B)learn the number of nucleotides contained within the genome.
C)identify genes and their positions within the genome.
D)learn how gene products interact to produce phenotypes.
A)establish the nucleotide sequence.
B)learn the number of nucleotides contained within the genome.
C)identify genes and their positions within the genome.
D)learn how gene products interact to produce phenotypes.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
18
In comparing DNA sequences from two different eukaryotic organisms,which do you expect to vary the most?
A)sequences that code for exons
B)sequences that code for introns
C)sequences that code for a polypeptide
D)sequences that regulate transcription
E)sequences that code for micro-RNAs
A)sequences that code for exons
B)sequences that code for introns
C)sequences that code for a polypeptide
D)sequences that regulate transcription
E)sequences that code for micro-RNAs

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
19
Although transposable elements and simple tandem repeats (STRs)are both repetitive DNAs,they differ in that
A)STRs occur within exons;transposable elements occur within introns.
B)STRs occur within introns;transposable elements occur within exons.
C)the repeated unit in STRs is clustered one after another;transposable element repeats are scattered throughout the genome.
D)the repeated unit in STRs is much larger than the repeated unit of transposable elements.
E)variation in STR repeat number comes from STR movement in the genome;variation in transposable element repeat number comes from errors in DNA replication.
A)STRs occur within exons;transposable elements occur within introns.
B)STRs occur within introns;transposable elements occur within exons.
C)the repeated unit in STRs is clustered one after another;transposable element repeats are scattered throughout the genome.
D)the repeated unit in STRs is much larger than the repeated unit of transposable elements.
E)variation in STR repeat number comes from STR movement in the genome;variation in transposable element repeat number comes from errors in DNA replication.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
20
Why are transposable elements considered to be selfish genes?
A)They produce products toxic to the host.
B)They replicate outside of the chromosome.
C)They replicate using the host's resources without direct benefit to the host.
D)They spread rapidly throughout the genome.
E)They produce products that prevent cooperation between cells.
A)They produce products toxic to the host.
B)They replicate outside of the chromosome.
C)They replicate using the host's resources without direct benefit to the host.
D)They spread rapidly throughout the genome.
E)They produce products that prevent cooperation between cells.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
21
Figure 20.1 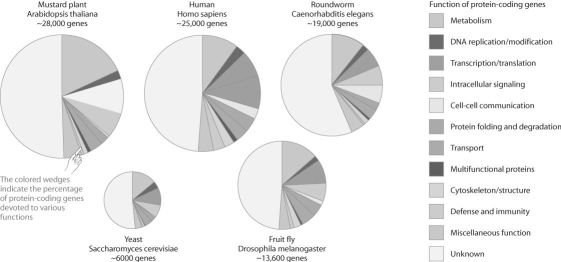
The figure above compares the proportion of genes devoted to various functions in a set of model organisms.Based on this figure,which one of the following statements is FALSE?
A)The function of roughly half of eukaryotic genes is unknown.
B)Humans have the highest proportion of genes devoted to defense and immunity.
C)The mustard plant and yeast have a small fraction of genes devoted to cell communication.
D)A relatively small but similar proportion of genes are devoted to transcription and translation in all these organisms.
E)Drosophila is unusual in having so few genes devoted to protein folding and degradation.

The figure above compares the proportion of genes devoted to various functions in a set of model organisms.Based on this figure,which one of the following statements is FALSE?
A)The function of roughly half of eukaryotic genes is unknown.
B)Humans have the highest proportion of genes devoted to defense and immunity.
C)The mustard plant and yeast have a small fraction of genes devoted to cell communication.
D)A relatively small but similar proportion of genes are devoted to transcription and translation in all these organisms.
E)Drosophila is unusual in having so few genes devoted to protein folding and degradation.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
22
If alternative splicing did not occur,a prediction would be that
A)the human genome would contain many more genes.
B)the yeast genome would contain many fewer genes.
C)there would be little correlation between the complexity of organisms and genome size.
D)there would be many fewer genes devoted to metabolism in Arabidopsis and yeast.
E)there would be many fewer genes devoted to DNA replication and modification in humans.
A)the human genome would contain many more genes.
B)the yeast genome would contain many fewer genes.
C)there would be little correlation between the complexity of organisms and genome size.
D)there would be many fewer genes devoted to metabolism in Arabidopsis and yeast.
E)there would be many fewer genes devoted to DNA replication and modification in humans.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
23
What can proteomics reveal that genomics cannot?
A)The number of genes characteristic of a species.
B)The patterns of alternative splicing.
C)The set of proteins present within a cell or tissue type.
D)The levels of mRNAs present in a particular cell type.
E)The movement of transposable elements within the genome.
A)The number of genes characteristic of a species.
B)The patterns of alternative splicing.
C)The set of proteins present within a cell or tissue type.
D)The levels of mRNAs present in a particular cell type.
E)The movement of transposable elements within the genome.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
24
If practical applications arise from this or similar metagenomic studies,what might these be?
A)New genes with useful properties may be identified and ultimately put to use.
B)New microbial species could be isolated from seawater.
C)An increased knowledge of the diversity of marine ecosystems could lead to increased appreciation of diversity in all ecosystems.
D)Species discovered in the Sargasso Sea may also be found to inhabit less-remote waters.
A)New genes with useful properties may be identified and ultimately put to use.
B)New microbial species could be isolated from seawater.
C)An increased knowledge of the diversity of marine ecosystems could lead to increased appreciation of diversity in all ecosystems.
D)Species discovered in the Sargasso Sea may also be found to inhabit less-remote waters.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
25
Mutations that occur in one member of a gene pair that arose from genes duplication may create
A)a pseudogene.
B)a gene with a new function.
C)a gene family with two distinct but related members.
D)A and B only.
E)all of the above.
A)a pseudogene.
B)a gene with a new function.
C)a gene family with two distinct but related members.
D)A and B only.
E)all of the above.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
26
When gene duplication occurs to its ultimate extent by doubling all genes in a genome,what has occurred?
A)pseudogene creation
B)creation of a gene cluster
C)creation of a polyploid
D)creation of an aneuploid
E)creation of a diploid
A)pseudogene creation
B)creation of a gene cluster
C)creation of a polyploid
D)creation of an aneuploid
E)creation of a diploid

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
27
Which of the following questions is asked in functional genomics research?
A)What is the number of Drosophila genes?
B)How does the GC content of human DNA vary across the genome?
C)What is the pattern of gene expression during mouse development?
D)How many introns exist in the human CFTR gene?
E)How closely related are the visual pigment genes of mouse and human?
A)What is the number of Drosophila genes?
B)How does the GC content of human DNA vary across the genome?
C)What is the pattern of gene expression during mouse development?
D)How many introns exist in the human CFTR gene?
E)How closely related are the visual pigment genes of mouse and human?

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
28
If one wished to test the hypothesis that humans and chimps differ due to differences in the expression of a large set of shared genes,the technique to use would be
A)Southern blotting.
B)PCR.
C)DNA sequencing.
D)protein-protein interaction assays.
E)DNA microarray analysis.
A)Southern blotting.
B)PCR.
C)DNA sequencing.
D)protein-protein interaction assays.
E)DNA microarray analysis.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
29
Because DNAs for sequencing were chosen at random with no way of knowing which organism they came from,how could genes be identified?
A)by searching for ORFs,especially those with translation start and stop sites
B)by searching for similarity between predicted ORFs and those in existing databases
C)by searching for exons bounded by consensus donor and acceptor splice sites (those that mark exon/intron boundaries)
D)A and B only
E)all of the above
A)by searching for ORFs,especially those with translation start and stop sites
B)by searching for similarity between predicted ORFs and those in existing databases
C)by searching for exons bounded by consensus donor and acceptor splice sites (those that mark exon/intron boundaries)
D)A and B only
E)all of the above

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
30
Figure 20.2 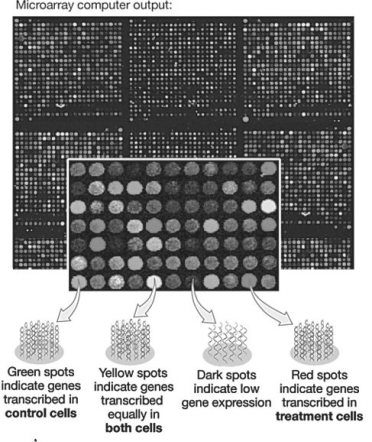
In the microarray shown in the figure above,if you were searching for genes whose expression was inhibited by hormone treatment,you would search for sequences spotted on the array that showed ________ fluorescence.
A)no
B)high levels of
C)green
D)red
E)yellow

In the microarray shown in the figure above,if you were searching for genes whose expression was inhibited by hormone treatment,you would search for sequences spotted on the array that showed ________ fluorescence.
A)no
B)high levels of
C)green
D)red
E)yellow

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
31
Like DNA microarrays,Northern blots analyze the levels of mRNA.However,Northern blots are almost always prepared with a probe tagged with one label,whereas DNA microarrays almost always use two groups of probes,each one tagged with a different fluorescent label.What advantage is conferred by using two labels for the microarray probes?
A)The two labels increase the sensitivity of the microarray.
B)The two labels allow the analysis of the expression of more genes.
C)The two labels allow simultaneous assay of levels of DNA and RNA.
D)The two labels allow comparison of gene expression between two samples.
E)Probing the array with two labels is equivalent to doing two experiments at once.
A)The two labels increase the sensitivity of the microarray.
B)The two labels allow the analysis of the expression of more genes.
C)The two labels allow simultaneous assay of levels of DNA and RNA.
D)The two labels allow comparison of gene expression between two samples.
E)Probing the array with two labels is equivalent to doing two experiments at once.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
32
Why was it necessary to modify the standard sequence assembly algorithms when analyzing DNAs from many different pooled species?
A)Sequencing techniques had improved sufficiently that obtaining accurate repetitive sequences was no longer a technical challenge.
B)Sequence-read lengths of repetitive DNA had increased sufficiently by the time of the Venter et al.work that they were almost as long as the sequences from nonrepetitive DNA.
C)DNAs from abundant organisms would occur frequently in the sample,making them look like repetitive DNAs.
D)DNAs from the largest organisms would occur infrequently in the sample,making them look like repetitive DNA.
A)Sequencing techniques had improved sufficiently that obtaining accurate repetitive sequences was no longer a technical challenge.
B)Sequence-read lengths of repetitive DNA had increased sufficiently by the time of the Venter et al.work that they were almost as long as the sequences from nonrepetitive DNA.
C)DNAs from abundant organisms would occur frequently in the sample,making them look like repetitive DNAs.
D)DNAs from the largest organisms would occur infrequently in the sample,making them look like repetitive DNA.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
33
Comparisons between bacterial pathogens and related nonpathogenic strains have revealed
A)additional genes in the pathogen that are associated with virulence.
B)a reduced numbers of genes in the pathogen that force it to adopt a parasitic lifestyle.
C)additional genes in the pathogen that free it from a parasitic lifestyle.
D)loss of genes in the pathogen that normally allow detection by the host's immune system.
A)additional genes in the pathogen that are associated with virulence.
B)a reduced numbers of genes in the pathogen that force it to adopt a parasitic lifestyle.
C)additional genes in the pathogen that free it from a parasitic lifestyle.
D)loss of genes in the pathogen that normally allow detection by the host's immune system.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
34
Table 20.1 The following table from the Venter et al. paper presents the numbers of different types of genes discovered in this study.
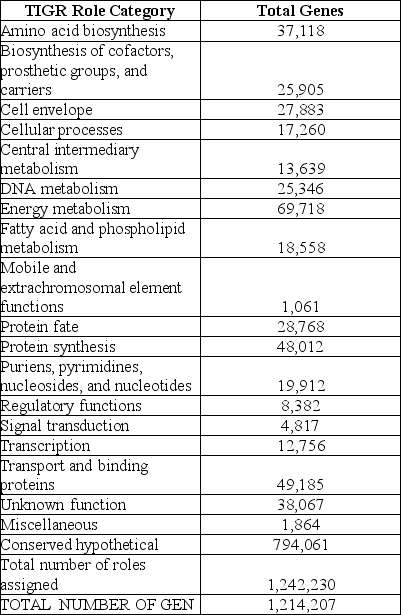
How do you think putative genes were placed into the categories shown in the preceding table?
A)by searching for ORFs,especially those with translation start and stop sites
B)by searching for matches,especially of the predicted gene products,with previously identified gene products of known function
C)by expressing the cloned genes in E.coli and then running biochemical analyses of the products
D)all of the above

How do you think putative genes were placed into the categories shown in the preceding table?
A)by searching for ORFs,especially those with translation start and stop sites
B)by searching for matches,especially of the predicted gene products,with previously identified gene products of known function
C)by expressing the cloned genes in E.coli and then running biochemical analyses of the products
D)all of the above

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
35
What is the main goal of obtaining a detailed human haplotype map?
A)to develop more effective vaccines
B)to locate introns
C)to locate regulatory elements
D)to locate genes associated with disease
E)to locate virulence genes
A)to develop more effective vaccines
B)to locate introns
C)to locate regulatory elements
D)to locate genes associated with disease
E)to locate virulence genes

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
36
A DNA microarray is a tool that owes its existence to earlier genomics investigations.What essential contribution of genomics makes microarrays possible?
A)recently improved RNA sequencing technologies
B)continuously improving methods of gene cloning
C)more efficient techniques for cDNA synthesis
D)knowledge of which DNA sequences to synthesize for the array
E)The concept that hybridization between single-stranded nucleic acids can be used as a means of identifying any DNA sequence.
A)recently improved RNA sequencing technologies
B)continuously improving methods of gene cloning
C)more efficient techniques for cDNA synthesis
D)knowledge of which DNA sequences to synthesize for the array
E)The concept that hybridization between single-stranded nucleic acids can be used as a means of identifying any DNA sequence.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
37
A current view of how the human and chimpanzee can share most of their nucleotide sequence yet exhibit significant phenotypic differences is that many of the most important sequence differences alter
A)structural genes.
B)the number of repeated sequences.
C)regulatory sequences and proteins.
D)environmental factors.
E)cultural factors.
A)structural genes.
B)the number of repeated sequences.
C)regulatory sequences and proteins.
D)environmental factors.
E)cultural factors.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
38
In eukaryotes,the major mechanism of introducing new genes into the genome is
A)duplication and divergence.
B)lateral gene transfer.
C)pseudogene creation.
D)pseudogene restoration.
E)unequal crossing over at microsatellite repeats.
A)duplication and divergence.
B)lateral gene transfer.
C)pseudogene creation.
D)pseudogene restoration.
E)unequal crossing over at microsatellite repeats.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
39
The features that are mapped in the HapMap Project are
A)restriction enzyme recognition sites.
B)promoters.
C)enhancers.
D)single-nucleotide differences.
E)STS repeat differences.
A)restriction enzyme recognition sites.
B)promoters.
C)enhancers.
D)single-nucleotide differences.
E)STS repeat differences.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
40
Why would any sequence assembly program be written so as to ignore repeated sequences?
A)These sequences are too unreliable to consider.
B)These sequences are too hard to obtain to justify the effort.
C)These sequences are too short to be useful for assembly.
D)These sequences are difficult to place within an assembly.
A)These sequences are too unreliable to consider.
B)These sequences are too hard to obtain to justify the effort.
C)These sequences are too short to be useful for assembly.
D)These sequences are difficult to place within an assembly.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck
41
As with many new endeavors,concerns have been raised by some that genomics will fundamentally change the way biological research is practiced.Examine the full reference for the Venter et al.paper and consider the way this work was done.Now think of some seminal works in the history of biology,for example,Darwin's On the Origin of Species and Watson and Crick's publication on DNA structure.What do you think the concern might be that has been voiced about genomics?
A)Genomics will offer deep insights too rapidly for the biological community to comprehend.
B)Genomics is so based in technology that it cannot possibly reveal important knowledge of biology.
C)Genomics makes biology a "big-team science" that will squeeze out innovative individuals or small groups of scientists who traditionally have provided our greatest insights.
D)Genomics is really computer science,not biology,and therefore offers little for understanding life.
A)Genomics will offer deep insights too rapidly for the biological community to comprehend.
B)Genomics is so based in technology that it cannot possibly reveal important knowledge of biology.
C)Genomics makes biology a "big-team science" that will squeeze out innovative individuals or small groups of scientists who traditionally have provided our greatest insights.
D)Genomics is really computer science,not biology,and therefore offers little for understanding life.

Unlock Deck
Unlock for access to all 41 flashcards in this deck.
Unlock Deck
k this deck


