Deck 26: Phylogeny and the Tree of Life
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/90
Play
Full screen (f)
Deck 26: Phylogeny and the Tree of Life
1
Which of these illustrates the correct representation of the binomial scientific name for the African lion?
A)Panthera leo
B)panthera leo
C)Panthera leo
D)Panthera Leo
E)Panthera leo
A)Panthera leo
B)panthera leo
C)Panthera leo
D)Panthera Leo
E)Panthera leo
E
2
The common ancestors of birds and mammals were very early (stem)reptiles, which almost certainly possessed 3-chambered hearts (2 atria, 1 ventricle). Birds and mammals, however, are alike in having 4-chambered hearts (2 atria, 2 ventricles). The 4-chambered hearts of birds and mammals are best described as
A)structural homologies.
B)vestiges.
C)homoplasies.
D)the result of shared ancestry.
E)molecular homologies.
A)structural homologies.
B)vestiges.
C)homoplasies.
D)the result of shared ancestry.
E)molecular homologies.
C
3
In angiosperm plants, flower morphology can be very intricate. If a tree, such as a New Mexico locust, has flowers that share many morphological intricacies with flowers of the sweet pea vine, then the most likely explanation for these floral similarities is the same general explanation for the similarities between the
A)dorsal fins of sharks and of dolphins.
B)reduced eyes of Australian moles and North American moles.
C)scales on moth wings and the scales of fish skin.
D)cranial bones of humans and those of chimpanzees.
E)adaptations for flight in birds and adaptations for flight in bats.
A)dorsal fins of sharks and of dolphins.
B)reduced eyes of Australian moles and North American moles.
C)scales on moth wings and the scales of fish skin.
D)cranial bones of humans and those of chimpanzees.
E)adaptations for flight in birds and adaptations for flight in bats.
D
4
Which mutation should least require realignment of homologous regions of a gene that is common to several related species?
A)3-base insertion
B)1-base substitution
C)4-base insertion
D)1-base deletion
E)3-base deletion
A)3-base insertion
B)1-base substitution
C)4-base insertion
D)1-base deletion
E)3-base deletion

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
5
The term "homoplasy" is most applicable to which of these features?
A)the legless condition found in various types of extant lizards
B)the 5-digit condition of human hands and bat wings
C)the beta-hemoglobin genes of mice and of humans
D)the fur that covers Australian moles and North American moles
E)the basic skeletal features of dog forelimbs and cat forelimbs
A)the legless condition found in various types of extant lizards
B)the 5-digit condition of human hands and bat wings
C)the beta-hemoglobin genes of mice and of humans
D)the fur that covers Australian moles and North American moles
E)the basic skeletal features of dog forelimbs and cat forelimbs

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
6
If organisms A, B, and C belong to the same class but to different orders and if organisms D, E, and F belong to the same order but to different families, which of the following pairs of organisms would be expected to show the greatest degree of structural homology?
A)A and B
B)A and C
C)B and D
D)C and F
E)D and F
A)A and B
B)A and C
C)B and D
D)C and F
E)D and F

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
7
The importance of computers and of computer software to modern cladistics is most closely linked to advances in
A)light microscopy.
B)radiometric dating.
C)fossil discovery techniques.
D)Linnaean classification.
E)molecular genetics.
A)light microscopy.
B)radiometric dating.
C)fossil discovery techniques.
D)Linnaean classification.
E)molecular genetics.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
8
Generally, within a lineage, the largest number of shared derived characters should be found among two organisms that are members of the same
A)kingdom.
B)class.
C)domain.
D)family.
E)order.
A)kingdom.
B)class.
C)domain.
D)family.
E)order.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
9
The legless condition that is observed in several groups of extant reptiles is the result of
A)their common ancestor having been legless.
B)a shared adaptation to an arboreal (living in trees)lifestyle.
C)several instances of the legless condition arising independently of each other.
D)individual lizards adapting to a fossorial (living in burrows)lifestyle during their lifetimes.
A)their common ancestor having been legless.
B)a shared adaptation to an arboreal (living in trees)lifestyle.
C)several instances of the legless condition arising independently of each other.
D)individual lizards adapting to a fossorial (living in burrows)lifestyle during their lifetimes.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
10
Some molecular data place the giant panda in the bear family (Ursidae)but place the lesser panda in the raccoon family (Procyonidae). Consequently, the morphological similarities of these two species are probably due to
A)inheritance of acquired characteristics.
B)sexual selection.
C)inheritance of shared derived characters.
D)possession of analogous structures.
E)possession of shared primitive characters.
A)inheritance of acquired characteristics.
B)sexual selection.
C)inheritance of shared derived characters.
D)possession of analogous structures.
E)possession of shared primitive characters.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
11
The scientific discipline concerned with naming organisms is called
A)taxonomy.
B)cladistics.
C)binomial nomenclature.
D)systematics.
E)phylocode
A)taxonomy.
B)cladistics.
C)binomial nomenclature.
D)systematics.
E)phylocode

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
12
Which of the following pairs are the best examples of homologous structures?
A)bat wing and human hand
B)owl wing and hornet wing
C)porcupine quill and cactus spine
D)bat forelimb and bird wing
E)Australian mole and North American mole
A)bat wing and human hand
B)owl wing and hornet wing
C)porcupine quill and cactus spine
D)bat forelimb and bird wing
E)Australian mole and North American mole

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
13
Darwin analogized the effects of evolution as the above-ground portion of a many-branched tree, with extant species being the tips of the twigs. The common ancestor of two species is most analogous to which anatomical tree part?
A)a single twig that gets longer with time
B)a node where two twigs diverge
C)a twig that branches with time
D)the trunk
E)neighboring twigs attached to the same stem
A)a single twig that gets longer with time
B)a node where two twigs diverge
C)a twig that branches with time
D)the trunk
E)neighboring twigs attached to the same stem

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
14
The best classification system is that which most closely
A)unites organisms that possess similar morphologies.
B)conforms to traditional, Linnaean taxonomic practices.
C)reflects evolutionary history.
D)corroborates the classification scheme in use at the time of Charles Darwin.
E)reflects the basic separation of prokaryotes from eukaryotes.
A)unites organisms that possess similar morphologies.
B)conforms to traditional, Linnaean taxonomic practices.
C)reflects evolutionary history.
D)corroborates the classification scheme in use at the time of Charles Darwin.
E)reflects the basic separation of prokaryotes from eukaryotes.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
15
The correct sequence, from the most to the least comprehensive, of the taxonomic levels listed here is
A)family, phylum, class, kingdom, order, species, and genus.
B)kingdom, phylum, class, order, family, genus, and species.
C)kingdom, phylum, order, class, family, genus, and species.
D)phylum, kingdom, order, class, species, family, and genus.
E)phylum, family, class, order, kingdom, genus, and species.
A)family, phylum, class, kingdom, order, species, and genus.
B)kingdom, phylum, class, order, family, genus, and species.
C)kingdom, phylum, order, class, family, genus, and species.
D)phylum, kingdom, order, class, species, family, and genus.
E)phylum, family, class, order, kingdom, genus, and species.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
16
Dozens of potato varieties exist, differing from each other in potato-tuber size, skin color, flesh color, and shape. One might construct a classification of potatoes based on these morphological traits. Which of these criticisms of such a classification scheme is most likely to come from an adherent of the phylocode method of classification?
A)Flesh color, rather than skin color, is a valid trait to use for classification because it is less susceptible to change with the age of the tuber.
B)Flower color is a better classification criterion, because below-ground tubers can be influenced by minerals in the soil as much as by their genes.
C)A more useful classification would codify potatoes based on the texture and flavor of their flesh, because this is what humans are concerned with.
D)The most accurate phylogenetic code is that of Linnaeus. Classify potatoes based on Linnaean principles; not according to their color.
E)The only biologically valid classification of potato varieties is one that accurately reflects their genetic and evolutionary relatedness.
A)Flesh color, rather than skin color, is a valid trait to use for classification because it is less susceptible to change with the age of the tuber.
B)Flower color is a better classification criterion, because below-ground tubers can be influenced by minerals in the soil as much as by their genes.
C)A more useful classification would codify potatoes based on the texture and flavor of their flesh, because this is what humans are concerned with.
D)The most accurate phylogenetic code is that of Linnaeus. Classify potatoes based on Linnaean principles; not according to their color.
E)The only biologically valid classification of potato varieties is one that accurately reflects their genetic and evolutionary relatedness.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
17
The common housefly belongs to all of the following taxa. Assuming you had access to textbooks or other scientific literature, knowing which of the following should provide you with the most specific information about the common housefly?
A)order Diptera
B)family Muscidae
C)genus Musca
D)class Hexapoda
E)phylum Arthropoda
A)order Diptera
B)family Muscidae
C)genus Musca
D)class Hexapoda
E)phylum Arthropoda

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
18
A phylogenetic tree that is "rooted" is one
A)that extends back to the origin of life on Earth.
B)at whose base is located the common ancestor of all taxa depicted on that tree.
C)that illustrates the rampant gene swapping that occurred early in life's history.
D)that indicates our uncertainty about the evolutionary relationships of the taxa depicted on the tree.
E)with very few branch points.
A)that extends back to the origin of life on Earth.
B)at whose base is located the common ancestor of all taxa depicted on that tree.
C)that illustrates the rampant gene swapping that occurred early in life's history.
D)that indicates our uncertainty about the evolutionary relationships of the taxa depicted on the tree.
E)with very few branch points.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
19
If, someday, an archaean cell is discovered whose SSU-rRNA sequence is more similar to that of humans than the sequence of mouse SSU-rRNA is to that of humans, the best explanation for this apparent discrepancy would be
A)homology.
B)homoplasy.
C)common ancestry.
D)retro-evolution by humans.
E)co-evolution of humans and that archaean.
A)homology.
B)homoplasy.
C)common ancestry.
D)retro-evolution by humans.
E)co-evolution of humans and that archaean.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
20
The various taxonomic levels (viz, genera, classes, etc.)of the hierarchical classification system differ from each other on the basis of
A)how widely the organisms assigned to each are distributed throughout the environment.
B)the body sizes of the organisms assigned to each.
C)their inclusiveness.
D)the relative genome sizes of the organisms assigned to each.
E)morphological characters that are applicable to all organisms.
A)how widely the organisms assigned to each are distributed throughout the environment.
B)the body sizes of the organisms assigned to each.
C)their inclusiveness.
D)the relative genome sizes of the organisms assigned to each.
E)morphological characters that are applicable to all organisms.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
21
The following questions refer to the hypothetical patterns of taxonomic hierarchy shown in Figure 26.2.

Figure 26.2
Which of the following numbers represents a polyphyletic taxon?
A)2
B)3
C)4
D)5
E)more than one of these

Figure 26.2
Which of the following numbers represents a polyphyletic taxon?
A)2
B)3
C)4
D)5
E)more than one of these

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
22
Use Figure 26.1 to answer the following questions.
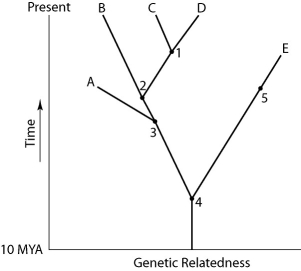
Figure 26.1
The two extant species that are most closely related to each other are
A)A and B.
B)B and C.
C)C and D.
D)D and E.
E)E and A.

Figure 26.1
The two extant species that are most closely related to each other are
A)A and B.
B)B and C.
C)C and D.
D)D and E.
E)E and A.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
23
The following questions refer to the hypothetical patterns of taxonomic hierarchy shown in Figure 26.2.

Figure 26.2
Shared derived characters are most likely to be found in taxa that are
A)paraphyletic.
B)polyphyletic.
C)monophyletic.

Figure 26.2
Shared derived characters are most likely to be found in taxa that are
A)paraphyletic.
B)polyphyletic.
C)monophyletic.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
24
The following questions refer to the hypothetical patterns of taxonomic hierarchy shown in Figure 26.2.

Figure 26.2
If the eukaryotic condition arose, independently, several different times during evolutionary history, and if ancestors of these different lineages are extant and are classified in the domain Eukarya, then the domain Eukarya would be
A)paraphyletic.
B)polyphyletic.
C)monophyletic.

Figure 26.2
If the eukaryotic condition arose, independently, several different times during evolutionary history, and if ancestors of these different lineages are extant and are classified in the domain Eukarya, then the domain Eukarya would be
A)paraphyletic.
B)polyphyletic.
C)monophyletic.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
25
Use Figure 26.1 to answer the following questions.

Figure 26.1
Which species are extinct?
A)A and E
B)A and B
C)C and D
D)D and E
E)cannot be determined from the information provided

Figure 26.1
Which species are extinct?
A)A and E
B)A and B
C)C and D
D)D and E
E)cannot be determined from the information provided

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
26
The following questions refer to the hypothetical patterns of taxonomic hierarchy shown in Figure 26.2.

Figure 26.2
The four-chambered hearts of birds and the four-chambered hearts of mammals evolved independently of each other. If one were unaware of this independence, then one might logically conclude that
A)the birds were the first to evolve a 4-chambered heart.
B)birds and mammals are more distantly related than is actually the case.
C)early mammals possessed feathers.
D)the common ancestor of birds and mammals had a four-chambered heart.
E)birds and mammals should be placed in the same family.

Figure 26.2
The four-chambered hearts of birds and the four-chambered hearts of mammals evolved independently of each other. If one were unaware of this independence, then one might logically conclude that
A)the birds were the first to evolve a 4-chambered heart.
B)birds and mammals are more distantly related than is actually the case.
C)early mammals possessed feathers.
D)the common ancestor of birds and mammals had a four-chambered heart.
E)birds and mammals should be placed in the same family.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
27
The following questions refer to the hypothetical patterns of taxonomic hierarchy shown in Figure 26.2.

Figure 26.2
Phylogenetic hypotheses (such as those represented by phylogenetic trees)are strongest when
A)they are based on amino acid sequences from homologous proteins, as long as the genes that code for such proteins contain no introns.
B)each clade is defined by a single derived character.
C)they are supported by more than one kind of evidence, such as when fossil evidence corroborates molecular evidence.
D)they are accepted by the foremost authorities in the field, especially if they have won Nobel Prizes.
E)they are based on a single DNA sequence that seems to be a shared derived sequence.

Figure 26.2
Phylogenetic hypotheses (such as those represented by phylogenetic trees)are strongest when
A)they are based on amino acid sequences from homologous proteins, as long as the genes that code for such proteins contain no introns.
B)each clade is defined by a single derived character.
C)they are supported by more than one kind of evidence, such as when fossil evidence corroborates molecular evidence.
D)they are accepted by the foremost authorities in the field, especially if they have won Nobel Prizes.
E)they are based on a single DNA sequence that seems to be a shared derived sequence.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
28
The following questions refer to the information below.
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.
-What conclusion can be drawn validly from these data?
A)Humans and other primates evolved from rabbits within the past 10 million years.
B)Most of the genes of other organisms are paralogous to human genes, or with chimpanzee genes.
C)Among the organisms listed, humans shared a common ancestor most recently with chimpanzees.
D)Humans evolved from chimpanzees somewhere in Africa within the last 6 million years.
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.
-What conclusion can be drawn validly from these data?
A)Humans and other primates evolved from rabbits within the past 10 million years.
B)Most of the genes of other organisms are paralogous to human genes, or with chimpanzee genes.
C)Among the organisms listed, humans shared a common ancestor most recently with chimpanzees.
D)Humans evolved from chimpanzees somewhere in Africa within the last 6 million years.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
29
The following questions refer to the hypothetical patterns of taxonomic hierarchy shown in Figure 26.2.

Figure 26.2
The term that is most appropriately associated with clade is
A)paraphyletic.
B)polyphyletic.
C)monophyletic.

Figure 26.2
The term that is most appropriately associated with clade is
A)paraphyletic.
B)polyphyletic.
C)monophyletic.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
30
The following questions refer to the hypothetical patterns of taxonomic hierarchy shown in Figure 26.2.

Figure 26.2
Ultimately, which of these serves as the basis for both the principle of maximum parsimony and the principle that shared complexity indicates homology rather than analogy?
A)the laws of thermodynamics
B)Boyle's law
C)the laws of probability
D)chaos theory
E)Hutchinson's law

Figure 26.2
Ultimately, which of these serves as the basis for both the principle of maximum parsimony and the principle that shared complexity indicates homology rather than analogy?
A)the laws of thermodynamics
B)Boyle's law
C)the laws of probability
D)chaos theory
E)Hutchinson's law

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
31
The following questions refer to the hypothetical patterns of taxonomic hierarchy shown in Figure 26.2.

Figure 26.2
Cladograms (a type of phylogenetic tree)constructed from evidence from molecular systematics are based on similarities in
A)morphology.
B)the pattern of embryological development.
C)biochemical pathways.
D)habitat and lifestyle choices.
E)mutations to homologous genes.

Figure 26.2
Cladograms (a type of phylogenetic tree)constructed from evidence from molecular systematics are based on similarities in
A)morphology.
B)the pattern of embryological development.
C)biochemical pathways.
D)habitat and lifestyle choices.
E)mutations to homologous genes.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
32
The following questions refer to the hypothetical patterns of taxonomic hierarchy shown in Figure 26.2.

Figure 26.2
Which of the following is not True of all horizontally oriented phylogenetic trees, where time advances to the right?
A)Each branch point represents a point in absolute time.
B)Organisms represented at the base of such trees are ancestral to those represented at higher levels.
C)The more branch points that occur between two taxa, the more divergent their DNA sequences should be.
D)The common ancestor represented by the rightmost branch point existed more recently in time than the common ancestors represented at branch points located to the left.
E)The more branch points there are, the more taxa are likely to be represented.

Figure 26.2
Which of the following is not True of all horizontally oriented phylogenetic trees, where time advances to the right?
A)Each branch point represents a point in absolute time.
B)Organisms represented at the base of such trees are ancestral to those represented at higher levels.
C)The more branch points that occur between two taxa, the more divergent their DNA sequences should be.
D)The common ancestor represented by the rightmost branch point existed more recently in time than the common ancestors represented at branch points located to the left.
E)The more branch points there are, the more taxa are likely to be represented.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
33
The following questions refer to the information below.
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.
-What probably explains the inclusion of rabbits in this research?
A)Their short generation time provides a ready source of DNA.
B)They possess all of the shared derived characters as do the other species listed.
C)They are the closest known relatives of rhesus monkeys.
D)They are the outgroup.
E)They are the most recent common ancestor of the primates.
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.
-What probably explains the inclusion of rabbits in this research?
A)Their short generation time provides a ready source of DNA.
B)They possess all of the shared derived characters as do the other species listed.
C)They are the closest known relatives of rhesus monkeys.
D)They are the outgroup.
E)They are the most recent common ancestor of the primates.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
34
The following questions refer to the hypothetical patterns of taxonomic hierarchy shown in Figure 26.2.

Figure 26.2
If this figure is an accurate depiction of relatedness, then which taxon is unacceptable, based on cladistics?
A)1
B)2
C)3
D)4
E)5

Figure 26.2
If this figure is an accurate depiction of relatedness, then which taxon is unacceptable, based on cladistics?
A)1
B)2
C)3
D)4
E)5

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
35
Use Figure 26.1 to answer the following questions.

Figure 26.1
If this evolutionary tree is an accurate depiction of relatedness, then which of the following should be correct?
1) The entire tree is based on maximum parsimony.
2) If all species depicted here make up a taxon, this taxon is monophyletic.
3) The last common ancestor of species B and C occurred more recently than the last common ancestor of species D and E.
4) Species A is the direct ancestor of both species B and species C.
5) The species present at position 3 is ancestral to C, D, and E.
A)2 and 5
B)1 and 3
C)3 and 4
D)2, 3, and 4
E)1, 2, and 3

Figure 26.1
If this evolutionary tree is an accurate depiction of relatedness, then which of the following should be correct?
1) The entire tree is based on maximum parsimony.
2) If all species depicted here make up a taxon, this taxon is monophyletic.
3) The last common ancestor of species B and C occurred more recently than the last common ancestor of species D and E.
4) Species A is the direct ancestor of both species B and species C.
5) The species present at position 3 is ancestral to C, D, and E.
A)2 and 5
B)1 and 3
C)3 and 4
D)2, 3, and 4
E)1, 2, and 3

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
36
The following questions refer to the hypothetical patterns of taxonomic hierarchy shown in Figure 26.2.

Figure 26.2
If birds are excluded from the class Reptilia, the term that consequently describes the class Reptilia is
A)paraphyletic.
B)polyphyletic.
C)monophyletic.

Figure 26.2
If birds are excluded from the class Reptilia, the term that consequently describes the class Reptilia is
A)paraphyletic.
B)polyphyletic.
C)monophyletic.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
37
The following questions refer to the hypothetical patterns of taxonomic hierarchy shown in Figure 26.2.

Figure 26.2
When using a cladistic approach to systematics, which of the following is considered most important for classification?
A)shared primitive characters
B)analogous primitive characters
C)shared derived characters
D)the number of homoplasies
E)overall phenotypic similarity

Figure 26.2
When using a cladistic approach to systematics, which of the following is considered most important for classification?
A)shared primitive characters
B)analogous primitive characters
C)shared derived characters
D)the number of homoplasies
E)overall phenotypic similarity

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
38
The following questions refer to the hypothetical patterns of taxonomic hierarchy shown in Figure 26.2.

Figure 26.2
A taxon, all of whose members have the same common ancestor, is
A)paraphyletic.
B)polyphyletic.
C)monophyletic.

Figure 26.2
A taxon, all of whose members have the same common ancestor, is
A)paraphyletic.
B)polyphyletic.
C)monophyletic.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
39
Use Figure 26.1 to answer the following questions.

Figure 26.1
Which extinct species should be the best candidate to serve as the outgroup for the clade whose common ancestor occurs at position 2?
A)A
B)B
C)C
D)D
E)E

Figure 26.1
Which extinct species should be the best candidate to serve as the outgroup for the clade whose common ancestor occurs at position 2?
A)A
B)B
C)C
D)D
E)E

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
40
Use Figure 26.1 to answer the following questions.

Figure 26.1
A common ancestor for both species C and E could be at position number
A)1)
B)2)
C)3)
D)4)
E)5)

Figure 26.1
A common ancestor for both species C and E could be at position number
A)1)
B)2)
C)3)
D)4)
E)5)

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
41
What is True of gene duplication (NOTE: gene duplication is a process that is distinct from DNA replication)?
A)It is a type of point mutation.
B)Its occurrence is limited to diploid species.
C)Its occurrence is limited to organisms without functional DNA-repair enzymes.
D)It is most similar in its effects to a deletion mutation.
E)It can increase the size of a genome over evolutionary time.
A)It is a type of point mutation.
B)Its occurrence is limited to diploid species.
C)Its occurrence is limited to organisms without functional DNA-repair enzymes.
D)It is most similar in its effects to a deletion mutation.
E)It can increase the size of a genome over evolutionary time.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
42
The following questions refer to the information below.
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.
-Which kind of DNA should provide the best molecular clock for gauging the evolutionary relatedness of several species whose common ancestor became extinct billions of years ago?
A)that coding for ribosomal RNA
B)intronic DNA belonging to a gene whose product performs a crucial function
C)paralogous DNA that has lost its function (i.e., no longer codes for functional gene product)
D)mitochondrial DNA
E)exonic DNA that codes for a non-crucial part of a polypeptide
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.
-Which kind of DNA should provide the best molecular clock for gauging the evolutionary relatedness of several species whose common ancestor became extinct billions of years ago?
A)that coding for ribosomal RNA
B)intronic DNA belonging to a gene whose product performs a crucial function
C)paralogous DNA that has lost its function (i.e., no longer codes for functional gene product)
D)mitochondrial DNA
E)exonic DNA that codes for a non-crucial part of a polypeptide

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
43
Theoretically, molecular clocks are to molecular phylogenies as radiometric dating is to phylogenies that are based on the
A)fossil record.
B)geographic distribution of extant species.
C)morphological similarities among extant species.
D)amino acid sequences of homologous polypeptides.
A)fossil record.
B)geographic distribution of extant species.
C)morphological similarities among extant species.
D)amino acid sequences of homologous polypeptides.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
44
The following questions refer to the information below.
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.
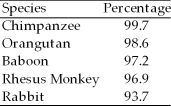
Figure 26.3
Typically, mutations that modify the active site of an enzyme are more likely to be harmful than mutations that affect other parts of the enzyme. A hypothetical enzyme consists of four domains (A-D), and the amino acid sequences of these four domains have been determined in five related species. Given the proportion of amino acid homologies among the five species at each of the four domains, which domain probably contains the active site?

A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.

Figure 26.3
Typically, mutations that modify the active site of an enzyme are more likely to be harmful than mutations that affect other parts of the enzyme. A hypothetical enzyme consists of four domains (A-D), and the amino acid sequences of these four domains have been determined in five related species. Given the proportion of amino acid homologies among the five species at each of the four domains, which domain probably contains the active site?


Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
45
The following questions refer to the information below.
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.
-Which statement represents the best explanation for the observation that the nuclear DNA of wolves and domestic dogs has a very high degree of homology?
A)Dogs and wolves have very similar morphologies.
B)Dogs and wolves belong to the same order.
C)Dogs and wolves are both members of the order Carnivora.
D)Dogs and wolves shared a common ancestor very recently.
E)Convergent evolution has occurred.
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.
-Which statement represents the best explanation for the observation that the nuclear DNA of wolves and domestic dogs has a very high degree of homology?
A)Dogs and wolves have very similar morphologies.
B)Dogs and wolves belong to the same order.
C)Dogs and wolves are both members of the order Carnivora.
D)Dogs and wolves shared a common ancestor very recently.
E)Convergent evolution has occurred.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
46
The following questions refer to the information below.
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.
-Concerning growth in genome size over evolutionary time, which of these does not belong with the others?
A)orthologous genes
B)gene duplications
C)paralogous genes
D)gene families
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.
-Concerning growth in genome size over evolutionary time, which of these does not belong with the others?
A)orthologous genes
B)gene duplications
C)paralogous genes
D)gene families

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
47
The following questions refer to the information below.
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.

Figure 26.3
When sufficient heat is applied, double-stranded DNA denatures into two single-stranded molecules as the heat breaks all of the hydrogen bonds. In an experiment, molecules of single-stranded DNA from species X are separately hybridized with putatively homologous single-stranded DNA molecules from five species (A-E). The hybridized DNAs are then heated, and the temperature at which complete denaturation occurs is recorded. Based on the data below, which species is probably most closely related to species X?

A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.

Figure 26.3
When sufficient heat is applied, double-stranded DNA denatures into two single-stranded molecules as the heat breaks all of the hydrogen bonds. In an experiment, molecules of single-stranded DNA from species X are separately hybridized with putatively homologous single-stranded DNA molecules from five species (A-E). The hybridized DNAs are then heated, and the temperature at which complete denaturation occurs is recorded. Based on the data below, which species is probably most closely related to species X?


Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
48
Neutral theory proposes that
A)molecular clocks are more reliable when the surrounding pH is close to 7.0.
B)most mutations of highly conserved DNA sequences should have no functional effect.
C)DNA is less susceptible to mutation when it codes for amino acid sequences whose side groups (or R groups)have a neutral pH.
D)DNA is less susceptible to mutation when it codes for amino acid sequences whose side groups (or R groups)have a neutral electrical charge.
E)a significant proportion of mutations is not acted upon by natural selection.
A)molecular clocks are more reliable when the surrounding pH is close to 7.0.
B)most mutations of highly conserved DNA sequences should have no functional effect.
C)DNA is less susceptible to mutation when it codes for amino acid sequences whose side groups (or R groups)have a neutral pH.
D)DNA is less susceptible to mutation when it codes for amino acid sequences whose side groups (or R groups)have a neutral electrical charge.
E)a significant proportion of mutations is not acted upon by natural selection.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
49
The following questions refer to the information below.
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.
-A researcher wants to determine the genetic relatedness of several breeds of dog (Canis familiaris). The researcher should compare homologous sequences of __________ that are known to be __________.
A)carbohydrates; poorly conserved
B)fatty acids; highly conserved
C)lipids; poorly conserved
D)proteins or nucleic acids; poorly conserved
E)amino acids; highly conserved
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.
-A researcher wants to determine the genetic relatedness of several breeds of dog (Canis familiaris). The researcher should compare homologous sequences of __________ that are known to be __________.
A)carbohydrates; poorly conserved
B)fatty acids; highly conserved
C)lipids; poorly conserved
D)proteins or nucleic acids; poorly conserved
E)amino acids; highly conserved

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
50
Morphologically, Species A is very similar to four other species, B-E. Yet the nucleotide sequence deep within an intron in a gene shared by all five of these eukaryotic species is quite different in Species A compared to that of the other four species when one studies the nucleotides present at each position.
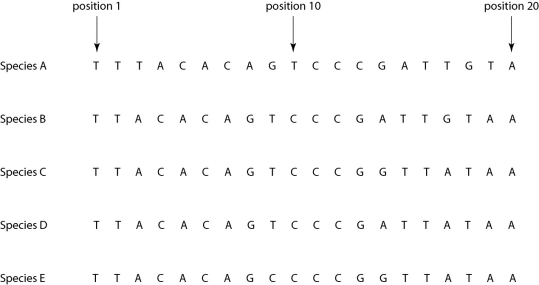 Figure 26.4
Figure 26.4
If the sequence of Species A differs from that of the other four species due to simple misalignment, then what should the computer software find when it compares the sequence of Species A to those of the other four species?
A)The nucleotide at position 1 should be different in Species A, but the same in species B-E.
B)The nucleotide sequence of Species A should have long sequences that are nearly identical to those of the other species, but offset in terms of position number.
C)The sequences of species B-E, though different from that of Species A, should be identical to each other, without exception.
D)If the software compares, not nucleotide sequence, but rather the amino acid sequence of the actual protein product, then the amino acid sequences of species B-E should be similar to each other, but very different from that of Species A.
E)Computer software is useless in determining sequences of introns; it can only be used with exons.
 Figure 26.4
Figure 26.4If the sequence of Species A differs from that of the other four species due to simple misalignment, then what should the computer software find when it compares the sequence of Species A to those of the other four species?
A)The nucleotide at position 1 should be different in Species A, but the same in species B-E.
B)The nucleotide sequence of Species A should have long sequences that are nearly identical to those of the other species, but offset in terms of position number.
C)The sequences of species B-E, though different from that of Species A, should be identical to each other, without exception.
D)If the software compares, not nucleotide sequence, but rather the amino acid sequence of the actual protein product, then the amino acid sequences of species B-E should be similar to each other, but very different from that of Species A.
E)Computer software is useless in determining sequences of introns; it can only be used with exons.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
51
The following questions refer to the information below.
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.
-Nucleic acid sequences that undergo few changes over the course of evolutionary time are said to be conserved. Conserved sequences of nucleic acids
A)are found in the most crucial portions of proteins.
B)include all mitochondrial DNA.
C)are abundant in ribosomes.
D)are proportionately more common in eukaryotic introns than in eukaryotic exons.
E)comprise a larger proportion of pre-mRNA (immature mRNA) than of mature mRNA.
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.
-Nucleic acid sequences that undergo few changes over the course of evolutionary time are said to be conserved. Conserved sequences of nucleic acids
A)are found in the most crucial portions of proteins.
B)include all mitochondrial DNA.
C)are abundant in ribosomes.
D)are proportionately more common in eukaryotic introns than in eukaryotic exons.
E)comprise a larger proportion of pre-mRNA (immature mRNA) than of mature mRNA.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
52
The following questions refer to the information below.
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.
-Species that are not closely related and that do not share many anatomical similarities can still be placed together on the same phylogenetic tree by comparing their
A)plasmids.
B)chloroplast genomes.
C)mitochondrial genomes.
D)homologous genes that are poorly conserved.
E)homologous genes that are highly conserved.
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.
-Species that are not closely related and that do not share many anatomical similarities can still be placed together on the same phylogenetic tree by comparing their
A)plasmids.
B)chloroplast genomes.
C)mitochondrial genomes.
D)homologous genes that are poorly conserved.
E)homologous genes that are highly conserved.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
53
The most important feature that permits a gene to act as a molecular clock is
A)having a large number of base pairs.
B)having a larger proportion of exonic DNA than of intronic DNA.
C)having a reliable average rate of mutation.
D)its recent origin by a gene-duplication event.
E)its being acted upon by natural selection.
A)having a large number of base pairs.
B)having a larger proportion of exonic DNA than of intronic DNA.
C)having a reliable average rate of mutation.
D)its recent origin by a gene-duplication event.
E)its being acted upon by natural selection.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
54
The following questions refer to the information below.
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.
-A phylogenetic tree constructed using sequence differences in mitochondrial DNA would be most valid for discerning the evolutionary relatedness of
A)archaeans and bacteria.
B)fungi and animals.
C)Hawaiian silverswords.
D)sharks and dolphins
E)mosses and ferns.
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.
-A phylogenetic tree constructed using sequence differences in mitochondrial DNA would be most valid for discerning the evolutionary relatedness of
A)archaeans and bacteria.
B)fungi and animals.
C)Hawaiian silverswords.
D)sharks and dolphins
E)mosses and ferns.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
55
The following questions refer to the information below.
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.
-If the genes of yeast are 50% orthologous to those of humans, and if the genes of mice are 99% orthologous to those of humans, then what percentage of the genes of fish might one validly predict to be orthologous to the genes of humans?
A)10%
B)30%
C)40%
D)50%
E)80%
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.
-If the genes of yeast are 50% orthologous to those of humans, and if the genes of mice are 99% orthologous to those of humans, then what percentage of the genes of fish might one validly predict to be orthologous to the genes of humans?
A)10%
B)30%
C)40%
D)50%
E)80%

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
56
The following questions refer to the information below.
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.
-Which of these items does not necessarily exist in a simple linear relationship with the number of gene-duplication events when placed as the label on the vertical axis of the graph below?
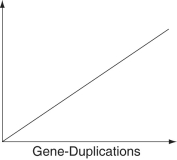
A)number of genes
B)number of DNA base pairs
C)genome size
D)mass (in picograms)of DNA
E)phenotypic complexity
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.
-Which of these items does not necessarily exist in a simple linear relationship with the number of gene-duplication events when placed as the label on the vertical axis of the graph below?

A)number of genes
B)number of DNA base pairs
C)genome size
D)mass (in picograms)of DNA
E)phenotypic complexity

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
57
The following questions refer to the information below.
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.
-The lakes of northern Minnesota are home to many similar species of damselflies of the genus Enallagma that have apparently undergone speciation from ancestral stock since the last glacial retreat about 10,000 years ago. Sequencing which of the following would probably be most useful in sorting out evolutionary relationships among these closely related species?
A)nuclear DNA
B)mitochondrial DNA
C)small nuclear RNA
D)ribosomal RNA
E)amino acids in proteins
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.
-The lakes of northern Minnesota are home to many similar species of damselflies of the genus Enallagma that have apparently undergone speciation from ancestral stock since the last glacial retreat about 10,000 years ago. Sequencing which of the following would probably be most useful in sorting out evolutionary relationships among these closely related species?
A)nuclear DNA
B)mitochondrial DNA
C)small nuclear RNA
D)ribosomal RNA
E)amino acids in proteins

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
58
The following questions refer to the information below.
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.
-The reason that paralogous genes can diverge from each other within the same gene pool, whereas orthologous genes diverge only after gene pools are isolated from each other, is that
A)having multiple copies of genes is essential for the occurrence of sympatric speciation in the wild.
B)paralogous genes can occur only in diploid species; thus, they are absent from most prokaryotes.
C)polyploidy is a necessary precondition for the occurrence of sympatric speciation in the wild.
D)having an extra copy of a gene permits modifications to the copy without loss of the original gene product.
A researcher compared the nucleotide sequences of a homologous gene from five different species of mammals with the homologous human gene. The sequence homology between each species' version of the gene and the human gene is presented as a percentage of similarity.
-The reason that paralogous genes can diverge from each other within the same gene pool, whereas orthologous genes diverge only after gene pools are isolated from each other, is that
A)having multiple copies of genes is essential for the occurrence of sympatric speciation in the wild.
B)paralogous genes can occur only in diploid species; thus, they are absent from most prokaryotes.
C)polyploidy is a necessary precondition for the occurrence of sympatric speciation in the wild.
D)having an extra copy of a gene permits modifications to the copy without loss of the original gene product.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
59
Paralogous genes that have lost the function of coding for a functional gene product are known as "pseudogenes." Which of these is a valid prediction regarding the fate of pseudogenes over evolutionary time?
A)They will be preserved by natural selection.
B)They will be highly conserved.
C)They will ultimately regain their original function.
D)They will be transformed into orthologous genes.
E)They will have relatively high mutation rates.
A)They will be preserved by natural selection.
B)They will be highly conserved.
C)They will ultimately regain their original function.
D)They will be transformed into orthologous genes.
E)They will have relatively high mutation rates.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
60
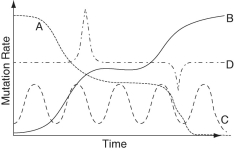
Which curve in the graph below best depicts the way that mutation rate varies over time in a gene that can serve as a reliable molecular clock?


Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
61
The following questions refer to this phylogenetic tree, depicting the origins of life and of the three domains. Horizontal lines indicate instances of gene or genome transfer.
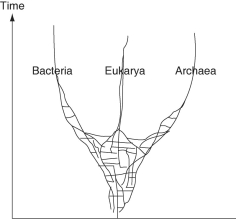
If the early history of life on Earth is accurately depicted by this phylogenetic tree, then which statement is least in agreement with the hypothesis proposed by this tree?
A)The last universal common ancestor of all extant species is better described as a community of organisms, rather than an individual species.
B)The origin of the three domains appears as a polytomy.
C)Archaean genomes should contain genes that originated in bacteria, and vice versa.
D)Eukaryotes are more closely related to archaeans than to bacteria.

If the early history of life on Earth is accurately depicted by this phylogenetic tree, then which statement is least in agreement with the hypothesis proposed by this tree?
A)The last universal common ancestor of all extant species is better described as a community of organisms, rather than an individual species.
B)The origin of the three domains appears as a polytomy.
C)Archaean genomes should contain genes that originated in bacteria, and vice versa.
D)Eukaryotes are more closely related to archaeans than to bacteria.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
62
The following questions refer to the table below, which compares the % sequence homology of four different parts (2 introns and 2 exons) of a gene that is found in five different eukaryotic species. Each part is numbered to indicate its distance from the promoter (e.g., Intron I is that closest to the promoter). The data reported for Species A were obtained by comparing DNA from one member of species A to another member of Species A.
-Which process hinders clarification of the deepest branchings in a phylogenetic tree that depicts the origins of the three domains?
A)binary fission
B)mitosis
C)meiosis
D)horizontal gene transfer
E)gene duplication
-Which process hinders clarification of the deepest branchings in a phylogenetic tree that depicts the origins of the three domains?
A)binary fission
B)mitosis
C)meiosis
D)horizontal gene transfer
E)gene duplication

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
63
Which of these would, if it had acted upon a gene, prevent this gene from acting as a reliable molecular clock?
A)neutral mutations
B)genetic drift
C)mutations within introns
D)natural selection
E)most substitution mutations involving an exonic codon's 3rd position
A)neutral mutations
B)genetic drift
C)mutations within introns
D)natural selection
E)most substitution mutations involving an exonic codon's 3rd position

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
64
The following questions refer to the table below, which compares the % sequence homology of four different parts (2 introns and 2 exons) of a gene that is found in five different eukaryotic species. Each part is numbered to indicate its distance from the promoter (e.g., Intron I is that closest to the promoter). The data reported for Species A were obtained by comparing DNA from one member of species A to another member of Species A.
-Which eukaryotic kingdom includes members that are the result of endosymbioses that included an ancient proteobacterium and an ancient cyanobacterium?
A)Plantae
B)Fungi
C)Animalia
D)Protista
E)Monera
-Which eukaryotic kingdom includes members that are the result of endosymbioses that included an ancient proteobacterium and an ancient cyanobacterium?
A)Plantae
B)Fungi
C)Animalia
D)Protista
E)Monera

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
65
The following questions refer to this phylogenetic tree, depicting the origins of life and of the three domains. Horizontal lines indicate instances of gene or genome transfer.

Which process is observed in prokaryotes and is responsible for the vertical components of the various bacterial and archaean lineages?
A)mitosis
B)meiosis
C)sexual reproduction
D)binary fission

Which process is observed in prokaryotes and is responsible for the vertical components of the various bacterial and archaean lineages?
A)mitosis
B)meiosis
C)sexual reproduction
D)binary fission

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
66
The following questions refer to the table below, which compares the % sequence homology of four different parts (2 introns and 2 exons) of a gene that is found in five different eukaryotic species. Each part is numbered to indicate its distance from the promoter (e.g., Intron I is that closest to the promoter). The data reported for Species A were obtained by comparing DNA from one member of species A to another member of Species A.
-Members of which kingdom have cell walls and are all heterotrophic?
A)Plantae
B)Fungi
C)Animalia
D)Protista
E)Monera
-Members of which kingdom have cell walls and are all heterotrophic?
A)Plantae
B)Fungi
C)Animalia
D)Protista
E)Monera

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
67
The following questions refer to the table below, which compares the % sequence homology of four different parts (2 introns and 2 exons) of a gene that is found in five different eukaryotic species. Each part is numbered to indicate its distance from the promoter (e.g., Intron I is that closest to the promoter). The data reported for Species A were obtained by comparing DNA from one member of species A to another member of Species A.
-Which kingdom has been replaced with two domains?
A)Plantae
B)Fungi
C)Animalia
D)Protista
E)Monera
-Which kingdom has been replaced with two domains?
A)Plantae
B)Fungi
C)Animalia
D)Protista
E)Monera

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
68
The following questions refer to the table below, which compares the % sequence homology of four different parts (2 introns and 2 exons) of a gene that is found in five different eukaryotic species. Each part is numbered to indicate its distance from the promoter (e.g., Intron I is that closest to the promoter). The data reported for Species A were obtained by comparing DNA from one member of species A to another member of Species A.
-Which of these four gene parts should allow the construction of the most accurate phylogenetic tree, assuming that this is the only part of the gene that has acted as a reliable molecular clock?
A)Intron I
B)Exon I
C)Intron VI
D)Exon V
-Which of these four gene parts should allow the construction of the most accurate phylogenetic tree, assuming that this is the only part of the gene that has acted as a reliable molecular clock?
A)Intron I
B)Exon I
C)Intron VI
D)Exon V

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
69
The following questions refer to the table below, which compares the % sequence homology of four different parts (2 introns and 2 exons) of a gene that is found in five different eukaryotic species. Each part is numbered to indicate its distance from the promoter (e.g., Intron I is that closest to the promoter). The data reported for Species A were obtained by comparing DNA from one member of species A to another member of Species A.
-What important criterion was used in the late 1960s to distinguish between the three multicellular eukaryotic kingdoms of the five-kingdom classification system?
A)the number of cells present in individual organisms
B)the geological stratum in which fossils first appear
C)the nutritional modes they employ
D)the biogeographic province where each first appears
E)the features of their embryos
-What important criterion was used in the late 1960s to distinguish between the three multicellular eukaryotic kingdoms of the five-kingdom classification system?
A)the number of cells present in individual organisms
B)the geological stratum in which fossils first appear
C)the nutritional modes they employ
D)the biogeographic province where each first appears
E)the features of their embryos

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
70
The following questions refer to the table below, which compares the % sequence homology of four different parts (2 introns and 2 exons) of a gene that is found in five different eukaryotic species. Each part is numbered to indicate its distance from the promoter (e.g., Intron I is that closest to the promoter). The data reported for Species A were obtained by comparing DNA from one member of species A to another member of Species A.
-Which of these is the best explanation for Intron I's relatively high sequence homology among these five species?
A)It is the most-upstream of this gene's introns.
B)It was once an exon, but became intronic in the common ancestor of these five species.
C)Due to alternative gene splicing, it is often treated as an exon in these five species; as an exon, it codes for an important part of a polypeptide.
D)It has a relatively high average rate of mutation.
-Which of these is the best explanation for Intron I's relatively high sequence homology among these five species?
A)It is the most-upstream of this gene's introns.
B)It was once an exon, but became intronic in the common ancestor of these five species.
C)Due to alternative gene splicing, it is often treated as an exon in these five species; as an exon, it codes for an important part of a polypeptide.
D)It has a relatively high average rate of mutation.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
71
The HIV genome's reliably high rate of change permits it to serve as a molecular clock. Which of these features is most responsible for this genome's high rate of change?
A)the relatively low number of nucleotides in the genome
B)the relatively small number of genes in the genome
C)the genome's ability to insert itself into the genome of the host
D)the lack of proofreading by the enzyme that converts HIV's RNA genome into a DNA genome
A)the relatively low number of nucleotides in the genome
B)the relatively small number of genes in the genome
C)the genome's ability to insert itself into the genome of the host
D)the lack of proofreading by the enzyme that converts HIV's RNA genome into a DNA genome

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
72
The following questions refer to the table below, which compares the % sequence homology of four different parts (2 introns and 2 exons) of a gene that is found in five different eukaryotic species. Each part is numbered to indicate its distance from the promoter (e.g., Intron I is that closest to the promoter). The data reported for Species A were obtained by comparing DNA from one member of species A to another member of Species A.
-Which of these is the best explanation for the relatively low level of sequence homology observed in Intron VI?
A)Mutations that occur here are neutral; thus, are neither selected for nor against, and thereby accumulate over time.
B)Its higher mutation rate has resulted in its highly conserved nature.
C)The occurrence of molecular homoplasy explains it.
D)This intron is not actually homologous, having resulted from separate bacteriophage-induced transduction events in these five species.
-Which of these is the best explanation for the relatively low level of sequence homology observed in Intron VI?
A)Mutations that occur here are neutral; thus, are neither selected for nor against, and thereby accumulate over time.
B)Its higher mutation rate has resulted in its highly conserved nature.
C)The occurrence of molecular homoplasy explains it.
D)This intron is not actually homologous, having resulted from separate bacteriophage-induced transduction events in these five species.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
73
The following questions refer to the table below, which compares the % sequence homology of four different parts (2 introns and 2 exons) of a gene that is found in five different eukaryotic species. Each part is numbered to indicate its distance from the promoter (e.g., Intron I is that closest to the promoter). The data reported for Species A were obtained by comparing DNA from one member of species A to another member of Species A.
-Regarding these sequence homology data, the principle of maximum parsimony would be applicable in
A)distinguishing introns from exons.
B)determining degree of sequence homology.
C)selecting appropriate genes for comparison among species.
D)inferring evolutionary relatedness from the number of sequence differences.
-Regarding these sequence homology data, the principle of maximum parsimony would be applicable in
A)distinguishing introns from exons.
B)determining degree of sequence homology.
C)selecting appropriate genes for comparison among species.
D)inferring evolutionary relatedness from the number of sequence differences.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
74
When it acts upon a gene, which of these processes consequently makes that gene an accurate molecular clock?
A)transcription
B)directional natural selection
C)mutation
D)proofreading
E)reverse transcription
A)transcription
B)directional natural selection
C)mutation
D)proofreading
E)reverse transcription

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
75
The following questions refer to the table below, which compares the % sequence homology of four different parts (2 introns and 2 exons) of a gene that is found in five different eukaryotic species. Each part is numbered to indicate its distance from the promoter (e.g., Intron I is that closest to the promoter). The data reported for Species A were obtained by comparing DNA from one member of species A to another member of Species A.
-Which is an obsolete kingdom that includes prokaryotic organisms?
A)Plantae
B)Fungi
C)Animalia
D)Protista
E)Monera
-Which is an obsolete kingdom that includes prokaryotic organisms?
A)Plantae
B)Fungi
C)Animalia
D)Protista
E)Monera

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
76
The following questions refer to the table below, which compares the % sequence homology of four different parts (2 introns and 2 exons) of a gene that is found in five different eukaryotic species. Each part is numbered to indicate its distance from the promoter (e.g., Intron I is that closest to the promoter). The data reported for Species A were obtained by comparing DNA from one member of species A to another member of Species A.
-What kind of evidence has recently made it necessary to assign the prokaryotes to either of two different domains, rather than assigning all prokaryotes to the same kingdom?
A)molecular
B)behavioral
C)nutritional
D)anatomical
E)ecological
-What kind of evidence has recently made it necessary to assign the prokaryotes to either of two different domains, rather than assigning all prokaryotes to the same kingdom?
A)molecular
B)behavioral
C)nutritional
D)anatomical
E)ecological

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
77
The following questions refer to the table below, which compares the % sequence homology of four different parts (2 introns and 2 exons) of a gene that is found in five different eukaryotic species. Each part is numbered to indicate its distance from the promoter (e.g., Intron I is that closest to the promoter). The data reported for Species A were obtained by comparing DNA from one member of species A to another member of Species A.
-The human nuclear genome includes hundreds of genes that are orthologs of bacterial genes, and hundreds of other genes that are orthologs of archaean genes. This finding can be explained by proposing that
A)neither archaea nor bacteria contain paralogous genes.
B)the eukaryotic lineage leading to humans involved at least one fusion of an ancient bacterium with an ancient archaean.
C)the infection of humans by bacteriophage introduced prokaryotic genes into the human genome.
D)horizontal gene transfer did not occur to any significant extent among the prokaryotic ancestors of humans.
-The human nuclear genome includes hundreds of genes that are orthologs of bacterial genes, and hundreds of other genes that are orthologs of archaean genes. This finding can be explained by proposing that
A)neither archaea nor bacteria contain paralogous genes.
B)the eukaryotic lineage leading to humans involved at least one fusion of an ancient bacterium with an ancient archaean.
C)the infection of humans by bacteriophage introduced prokaryotic genes into the human genome.
D)horizontal gene transfer did not occur to any significant extent among the prokaryotic ancestors of humans.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
78
The following questions refer to the table below, which compares the % sequence homology of four different parts (2 introns and 2 exons) of a gene that is found in five different eukaryotic species. Each part is numbered to indicate its distance from the promoter (e.g., Intron I is that closest to the promoter). The data reported for Species A were obtained by comparing DNA from one member of species A to another member of Species A.
-Based on the tabular data, and assuming that time advances vertically, which cladogram (a type of phylogenetic tree)is the most likely depiction of the evolutionary relationships among these five species?
A)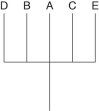
B)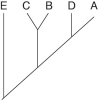
C)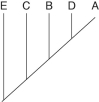
D)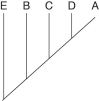
-Based on the tabular data, and assuming that time advances vertically, which cladogram (a type of phylogenetic tree)is the most likely depiction of the evolutionary relationships among these five species?
A)

B)

C)

D)


Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
79
The following questions refer to the table below, which compares the % sequence homology of four different parts (2 introns and 2 exons) of a gene that is found in five different eukaryotic species. Each part is numbered to indicate its distance from the promoter (e.g., Intron I is that closest to the promoter). The data reported for Species A were obtained by comparing DNA from one member of species A to another member of Species A.
-Which of these is the best explanation for the high degree of sequence homology observed in Exon I among these five species?
A)It is the most-upstream exon of this gene.
B)Due to alternative gene splicing, this exon is often treated as an intron.
C)It codes for a polypeptide domain that has a crucial function.
D)These five species must actually constitute a single species.
E)This exon is rich in G-C base pairs; thus, is more stable.
-Which of these is the best explanation for the high degree of sequence homology observed in Exon I among these five species?
A)It is the most-upstream exon of this gene.
B)Due to alternative gene splicing, this exon is often treated as an intron.
C)It codes for a polypeptide domain that has a crucial function.
D)These five species must actually constitute a single species.
E)This exon is rich in G-C base pairs; thus, is more stable.

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck
80
The following questions refer to the table below, which compares the % sequence homology of four different parts (2 introns and 2 exons) of a gene that is found in five different eukaryotic species. Each part is numbered to indicate its distance from the promoter (e.g., Intron I is that closest to the promoter). The data reported for Species A were obtained by comparing DNA from one member of species A to another member of Species A.
-Which eukaryotic kingdom is polyphyletic and therefore not acceptable, based on cladistics?
A)Plantae
B)Fungi
C)Animalia
D)Protista
E)Monera
-Which eukaryotic kingdom is polyphyletic and therefore not acceptable, based on cladistics?
A)Plantae
B)Fungi
C)Animalia
D)Protista
E)Monera

Unlock Deck
Unlock for access to all 90 flashcards in this deck.
Unlock Deck
k this deck


