Deck 10: Gene Isolation and Manipulation
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/55
Play
Full screen (f)
Deck 10: Gene Isolation and Manipulation
1
Which of the following statements about PCR is/are FALSE?
A)PCR reaction contains DNA template,primers,deoxyribonucleotide triphosphates,and heat-resistant DNA polymerase.
B)Each PCR cycle includes three steps: denaturation,annealing,and extension.
C)After each PCR cycle,amount of DNA increases in a linear manner.
D)One limitation in PCR is that it requires prior knowledge of the sequence or at least part of the sequence to be amplified.
E)PCR is a very sensitive technique and can amplify target sequences present in extremely low copy numbers.
A)PCR reaction contains DNA template,primers,deoxyribonucleotide triphosphates,and heat-resistant DNA polymerase.
B)Each PCR cycle includes three steps: denaturation,annealing,and extension.
C)After each PCR cycle,amount of DNA increases in a linear manner.
D)One limitation in PCR is that it requires prior knowledge of the sequence or at least part of the sequence to be amplified.
E)PCR is a very sensitive technique and can amplify target sequences present in extremely low copy numbers.
C
2
The polymerase chain reaction requires ssDNA primers that:
A)anneal to the same strand of template DNA,though at distant sites.
B)anneal to opposite strands of template DNA at distant sites,with their 3′ ends directed toward each other.
C)anneal to opposite strands of template DNA at distant sites,with their 5′ ends directed toward each other.
D)anneal to each other to prime DNA polymerization.
E)hybridize only to DNA within the open reading frame of a selected gene.
A)anneal to the same strand of template DNA,though at distant sites.
B)anneal to opposite strands of template DNA at distant sites,with their 3′ ends directed toward each other.
C)anneal to opposite strands of template DNA at distant sites,with their 5′ ends directed toward each other.
D)anneal to each other to prime DNA polymerization.
E)hybridize only to DNA within the open reading frame of a selected gene.
B
3
Which of the following is NOT true of a cDNA clone?
A)cDNAs are generally copies of mRNA from expressed genes.
B)Reverse transcriptase activity is required for cDNA cloning.
C)cDNA clones lack exons,as introns are spliced together before cloning.
D)cDNAs are typically smaller than the chromosomal sequence for a particular gene.
E)cDNAs can be fused to a promoter to enable expression of the encoded gene.
A)cDNAs are generally copies of mRNA from expressed genes.
B)Reverse transcriptase activity is required for cDNA cloning.
C)cDNA clones lack exons,as introns are spliced together before cloning.
D)cDNAs are typically smaller than the chromosomal sequence for a particular gene.
E)cDNAs can be fused to a promoter to enable expression of the encoded gene.
C
4
Which of the following sequences includes a clear eight base pair palindrome?
A)5′-CCGATCGATCCC-3′
B)5′-TGGGGTTTT G-3′
C)5′-GGGGAAAA-3′
D)5′-GATCCTAG-3′
E)5′-AACCAACCAA-3′
A)5′-CCGATCGATCCC-3′
B)5′-TGGGGTTTT G-3′
C)5′-GGGGAAAA-3′
D)5′-GATCCTAG-3′
E)5′-AACCAACCAA-3′

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
5
Gel electrophoresis can be used to separate DNA or RNA molecules based on their:
A)charge.
B)molecular weight.
C)solubility in buffer.
D)All of the answer options are correct.
E)charge and molecular weight.
A)charge.
B)molecular weight.
C)solubility in buffer.
D)All of the answer options are correct.
E)charge and molecular weight.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
6
A radioactive probe is generated using the actin gene from yeast.This probe is used in a Southern blot analysis of EcoRI digested genomic DNA from the ciliated protozoan Tetrahymena thermophila.The autoradiogram shows a single-labeled band of 4 kb in size.This means that the Tetrahymena actin gene is:
A)4 kb in size.
B)the same size as the yeast gene.
C)present in one copy in the genome.
D)present in four copies in the genome.
E)identical in sequence with the yeast gene.
A)4 kb in size.
B)the same size as the yeast gene.
C)present in one copy in the genome.
D)present in four copies in the genome.
E)identical in sequence with the yeast gene.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
7
In the Sanger sequencing method,the use of dideoxy adenosine triphosphate stops nucleotide polymerization opposite:
A)A's in the template strand.
B)T's in the template strand.
C)G's in the template strand.
D)C's in the template strand.
E)any base selected randomly in the template strand.
A)A's in the template strand.
B)T's in the template strand.
C)G's in the template strand.
D)C's in the template strand.
E)any base selected randomly in the template strand.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
8
Recombinant DNA techniques typically require the action of:
A)DNA polymerase and phosphatase.
B)restriction enzymes and DNA ligase.
C)RNA polymerase and RNA primase.
D)reverse transcriptase.
E)DNA helicase.
A)DNA polymerase and phosphatase.
B)restriction enzymes and DNA ligase.
C)RNA polymerase and RNA primase.
D)reverse transcriptase.
E)DNA helicase.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
9
A plasmid vector has a gene for erythromycin resistance (EryR)and a gene for ampicillin resistance (AmpR).The Amp gene is cut with restriction enzyme,and donor DNA treated with the same enzyme is added.What genotype of cells needs to be selected to show evidence of transformation?
A)AmpREryR
B)AmpREryS
C)AmpSEryR
D)AmpSEryS
E)None of the answer options are correct.
A)AmpREryR
B)AmpREryS
C)AmpSEryR
D)AmpSEryS
E)None of the answer options are correct.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
10
The cDNA for a eukaryotic gene B is 900 nucleotide pairs long.A cDNA clone is used to isolate a genomic clone of gene B,and the gene is sequenced.From start to stop codon,the gene is found to be 1800 nucleotide pairs long.The most probable reason for the discrepancy is that:
A)the mRNA broke during cDNA synthesis.
B)the gene is present as a tandem duplication.
C)the gene has 900 nucleotide pairs of introns.
D)the genomic clone is not really gene B,just a related gene.
E)there was a sequencing error.
A)the mRNA broke during cDNA synthesis.
B)the gene is present as a tandem duplication.
C)the gene has 900 nucleotide pairs of introns.
D)the genomic clone is not really gene B,just a related gene.
E)there was a sequencing error.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
11
Assume that a cosmid will carry inserts of about 50 kb and that cosmids are used to clone a 3 Mb (megabase)genome.Assuming you are particularly lucky and have no duplication in your library,what is the smallest number of cosmid clones you would need for a "complete" genomic library?
A)3000 clones
B)600 clones
C)300 clones
D)60 clones
E)30 clones
A)3000 clones
B)600 clones
C)300 clones
D)60 clones
E)30 clones

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
12
Identification of mRNA and initiation of priming for cDNA synthesis is accomplished by:
A)purifying only cytosolic RNAs before initiating the process.
B)coupling cDNA synthesis to exon splicing.
C)detecting the 5′ cap sequence to initiation cDNA synthesis.
D)use of oligo-dT to prime cDNA synthesis from the polyA tail.
E)column purification of mRNA sequences.
A)purifying only cytosolic RNAs before initiating the process.
B)coupling cDNA synthesis to exon splicing.
C)detecting the 5′ cap sequence to initiation cDNA synthesis.
D)use of oligo-dT to prime cDNA synthesis from the polyA tail.
E)column purification of mRNA sequences.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
13
Perhaps the most common use of plasmid vectors is to:
A)enable the amplification of cloned DNA within a bacterial host cell.
B)isolate viral DNA from infected cells.
C)insert restriction enzyme cut sites into cDNAs.
D)generate recombinant proteins.
E)manipulate the cell biology of bacterial cells.
A)enable the amplification of cloned DNA within a bacterial host cell.
B)isolate viral DNA from infected cells.
C)insert restriction enzyme cut sites into cDNAs.
D)generate recombinant proteins.
E)manipulate the cell biology of bacterial cells.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
14
A wild-type Aspergillus strain is transformed with a plasmid carrying a hygromycin resistance allele,and cells are plated on hygromycin.One resistant colony showed an aberrant type of aerial hyphae.When crossed to wild type,the progeny were 1/2 normal hyphae,Hyg sensitive,and 1/2 aberrant hyphae,Hyg-resistant.The probable explanation is that:
A)the plasmid was inserted in a gene for normal hyphal development.
B)the plasmid interfered with hyphal development.
C)a mutation arose in a gene for hyphal development on the plasmid.
D)a mutation arose in a gene for hyphal development on a recipient chromosome.
E)the recipient was a heterokaryon carrying some mutant nuclei.
A)the plasmid was inserted in a gene for normal hyphal development.
B)the plasmid interfered with hyphal development.
C)a mutation arose in a gene for hyphal development on the plasmid.
D)a mutation arose in a gene for hyphal development on a recipient chromosome.
E)the recipient was a heterokaryon carrying some mutant nuclei.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
15
In recombinant DNA technology,DNA is most often cut using:
A)DNA ligase.
B)DNA polymerase.
C)DNA gyrase.
D)restriction endonucleases.
E)terminal transferase.
A)DNA ligase.
B)DNA polymerase.
C)DNA gyrase.
D)restriction endonucleases.
E)terminal transferase.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
16
A circular DNA molecule has n target sites for restriction enzyme EcoRI.How many fragments will be produced after complete digestion?
A)n - 1
B)n
C)n + 1
D)2n - 1
E)2n + 2
A)n - 1
B)n
C)n + 1
D)2n - 1
E)2n + 2

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
17
The intermediate temperature cycle (72°C)in the polymerase chain reaction enables:
A)hybridization between the template DNA and the PCR primers.
B)denaturation of the template DNA to enable primer access to matching sequence.
C)activation of the primers to being the amplification procedure.
D)activity of the thermostable DNA polymerase enzyme.
E)a reduction of contamination in the amplification reaction.
A)hybridization between the template DNA and the PCR primers.
B)denaturation of the template DNA to enable primer access to matching sequence.
C)activation of the primers to being the amplification procedure.
D)activity of the thermostable DNA polymerase enzyme.
E)a reduction of contamination in the amplification reaction.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
18
Which of the following about plasmid vectors is/are TRUE?
A)Plasmid vectors must have convenient restriction sites/polylinker sites.
B)Plasmid vectors must have an origin of replication.
C)Plasmid vectors must carry a selectable marker (drug resistance)for selection.
D)Plasmid vectors can accept DNA inserts as large as 10 kb to 15 kb.
E)All of the answer options are correct.
A)Plasmid vectors must have convenient restriction sites/polylinker sites.
B)Plasmid vectors must have an origin of replication.
C)Plasmid vectors must carry a selectable marker (drug resistance)for selection.
D)Plasmid vectors can accept DNA inserts as large as 10 kb to 15 kb.
E)All of the answer options are correct.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
19
Which of the following about the Sanger sequencing method is/are TRUE?
A)Sanger sequencing uses dideoxynucleotide triphosphates to terminate DNA polymerization.
B)Dideoxynucleotides lack 2′ and 3′ OH groups in the ribose sugar.
C)All four deoxynucleotide triphosphates are used in a Sanger sequencing reaction.
D)Fluorescent dyes can be used to automate Sanger sequencing.
E)All of the answer options are correct.
A)Sanger sequencing uses dideoxynucleotide triphosphates to terminate DNA polymerization.
B)Dideoxynucleotides lack 2′ and 3′ OH groups in the ribose sugar.
C)All four deoxynucleotide triphosphates are used in a Sanger sequencing reaction.
D)Fluorescent dyes can be used to automate Sanger sequencing.
E)All of the answer options are correct.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
20
A linear DNA molecule has n target sites for restriction enzyme EcoRI.How many fragments will be produced after complete digestion?
A)n - 1
B)n
C)n + 1
D)2n - 1
E)2n + 2
A)n - 1
B)n
C)n + 1
D)2n - 1
E)2n + 2

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
21
A transgenic mouse is different from a knockout mouse in that:
A)transgenic mice have DNA added to their genome.
B)transgenic mice do not have DNA added to their genome.
C)the transgene is inserted at a random (ectopic)site in the genome.
D)a knockout mouse is more healthy than a transgenic mouse.
E)the knockout mouse is sterile.
A)transgenic mice have DNA added to their genome.
B)transgenic mice do not have DNA added to their genome.
C)the transgene is inserted at a random (ectopic)site in the genome.
D)a knockout mouse is more healthy than a transgenic mouse.
E)the knockout mouse is sterile.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
22
Embryonic stem (ES)cells have the unique ability to:
A)form a mouse pup without any additional cells.
B)combine with other stem cells to generate a chimeric mouse pup.
C)grow in the presence of gancyclovir.
D)eliminate specific genes in response to chemical cues.
E)divide more rapidly than other cell types.
A)form a mouse pup without any additional cells.
B)combine with other stem cells to generate a chimeric mouse pup.
C)grow in the presence of gancyclovir.
D)eliminate specific genes in response to chemical cues.
E)divide more rapidly than other cell types.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
23
Thymidine kinase is valuable in the targeting of mouse genes because:
A)it enhances the efficiency of gene replacement.
B)it suppresses expression of the target gene.
C)it allows researchers to enrich cells for those that have undergone homologous recombination.
D)transgenic cells are resistant to gancyclovir.
E)transgenic cells are sensitive to gancyclovir.
A)it enhances the efficiency of gene replacement.
B)it suppresses expression of the target gene.
C)it allows researchers to enrich cells for those that have undergone homologous recombination.
D)transgenic cells are resistant to gancyclovir.
E)transgenic cells are sensitive to gancyclovir.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
24
In the study of genetics,a "library" denotes:
A)the information carried in a digital database,such as GenBank.
B)a collection of DNA fragments isolated from a particular group of cells (or tissue)representing the genetic content of those cells.
C)a complex collection of restriction enzymes gathered for the purpose of restriction enzyme mapping.
D)all the proteins present in a particular cell type,collected by protein extraction.
E)a collection of books that explore the topic of genetics.
A)the information carried in a digital database,such as GenBank.
B)a collection of DNA fragments isolated from a particular group of cells (or tissue)representing the genetic content of those cells.
C)a complex collection of restriction enzymes gathered for the purpose of restriction enzyme mapping.
D)all the proteins present in a particular cell type,collected by protein extraction.
E)a collection of books that explore the topic of genetics.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
25
What type of library would be most valuable in the isolation of promoter sequences found in front of a gene's transcriptional start site?
A)genomic DNA library
B)cDNA library
C)miRNA library
D)mutant genomic DNA library
E)repetitive DNA library
A)genomic DNA library
B)cDNA library
C)miRNA library
D)mutant genomic DNA library
E)repetitive DNA library

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
26
Which of the following organisms can be made transgenic by injection of DNA into the gonad,generating extrachromosomal arrays of the transgene in some egg cells?
A)mouse (Mus musculus)
B)yeast (Saccharomyces cerevisiae)
C)bacteria (Escherichia coli)
D)humans (Homo sapiens)
E)roundworm (Caenorhabditis elegans)
A)mouse (Mus musculus)
B)yeast (Saccharomyces cerevisiae)
C)bacteria (Escherichia coli)
D)humans (Homo sapiens)
E)roundworm (Caenorhabditis elegans)

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
27
Suppose you cut two different DNAs,
then ligate them together through their compatible sticky ends.Once joined,could you separate these two DNAs again with either restriction enzyme? Why or why not? (Each of these enzymes is cut between the first two nucleotides,at the 5? end of each strand of the palindrome.)
then ligate them together through their compatible sticky ends.Once joined,could you separate these two DNAs again with either restriction enzyme? Why or why not? (Each of these enzymes is cut between the first two nucleotides,at the 5? end of each strand of the palindrome.)

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
28
Restriction endonucleases BamH1,NheI,XbaI,and EcoRI make sequence-specific cuts in DNA at the following sequences:
BamHI: 5'-GGATCC-3'
NheI: 5'-GCTAGC-3'
XbaI: 5'-TCTAGA-3'
EcoRI: 5'-GAATTC-3'
All these enzymes break a phosphodiester bond between the first and second nucleotide (counting from the 5' end),leaving a 5' P and a 3' OH.
Could ligase covalently join two ends produced by the following enzymes? If so,would ligation regenerate one of the above restriction sites?
(1)two BamH1 ends?
(2)two XbaI ends?
(3)a BamH1 and an EcoRI end?
(4)an XbaI and an NheI end?
(5)a BamH1 and an NheI end?
BamHI: 5'-GGATCC-3'
NheI: 5'-GCTAGC-3'
XbaI: 5'-TCTAGA-3'
EcoRI: 5'-GAATTC-3'
All these enzymes break a phosphodiester bond between the first and second nucleotide (counting from the 5' end),leaving a 5' P and a 3' OH.
Could ligase covalently join two ends produced by the following enzymes? If so,would ligation regenerate one of the above restriction sites?
(1)two BamH1 ends?
(2)two XbaI ends?
(3)a BamH1 and an EcoRI end?
(4)an XbaI and an NheI end?
(5)a BamH1 and an NheI end?

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
29
Foreign DNA can be introduced into a cell using which of the following method/s?
A)transformation
B)projectile gun
C)injection
D)virus infection
E)All of the answer options are correct.
A)transformation
B)projectile gun
C)injection
D)virus infection
E)All of the answer options are correct.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
30
Transgenic plants can be generated using T-DNA plasmid carrying a gene of interest.To get the DNA into the plant cells,the researchers:
A)use a syringe needle to inject the DNA into pollen before fertilization.
B)use a syringe needle to inject the DNA into groups of cells in the plant's root tissue.
C)co-cultivate bacteria with T-DNA and plant cells,resulting in DNA transfer.
D)add a mild detergent to cultures of plant cells to open holes in the cell wall.
E)remove the plant cell's normal DNA and replace it with the T-DNA.
A)use a syringe needle to inject the DNA into pollen before fertilization.
B)use a syringe needle to inject the DNA into groups of cells in the plant's root tissue.
C)co-cultivate bacteria with T-DNA and plant cells,resulting in DNA transfer.
D)add a mild detergent to cultures of plant cells to open holes in the cell wall.
E)remove the plant cell's normal DNA and replace it with the T-DNA.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
31
Bacterial cells have a variety of restriction enzymes that can degrade the DNA of an attacking virus.How do these bacterial cells protect their own DNA from being degraded by the very restriction enzymes that are meant to help them?

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
32
A 3-kb supercoiled circular plasmid contains the restriction sites shown below.N and X represent NheI and XbaI sites,respectively;A through F mark six points on the plasmid.A preparation of plasmid DNA was separately digested to completion with XbaI alone,NheI alone,and a mixture of the two enzymes.The products were separated by agarose gel electrophoresis.Draw the predicted appearance of the gel,assuming that you ran the following in the various lanes. 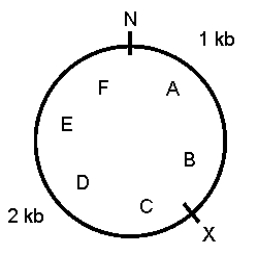
Lane M (size markers)-a mixture of known markers increasing in size from 1 kb to 6 kb,in steps of 1 kb
Lane 1-sample from the original plasmid DNA preparation
Lane 2-products of complete digestion with XbaI alone
Lane 3-products of complete digestion with NheI alone
Lane 4-products of complete digestion with XbaI plus NheI

Lane M (size markers)-a mixture of known markers increasing in size from 1 kb to 6 kb,in steps of 1 kb
Lane 1-sample from the original plasmid DNA preparation
Lane 2-products of complete digestion with XbaI alone
Lane 3-products of complete digestion with NheI alone
Lane 4-products of complete digestion with XbaI plus NheI

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
33
Homologous recombination in yeast facilitates the:
A)segregation of genes during meiosis.
B)targeted replacement of a gene in a living yeast cell.
C)amplification of homologous yeast genes.
D)independent assortment of separate gene alleles.
E)expression of similar gene sequences via one promoter.
A)segregation of genes during meiosis.
B)targeted replacement of a gene in a living yeast cell.
C)amplification of homologous yeast genes.
D)independent assortment of separate gene alleles.
E)expression of similar gene sequences via one promoter.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
34
This linear vector is comprised of bacteriophage genome components and can carry cloned DNA up to 15 kb in size.
A)pUC19
B)lambda
C)fosmid
D)bacterial artificial chromosome
E)T-vector
A)pUC19
B)lambda
C)fosmid
D)bacterial artificial chromosome
E)T-vector

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
35
Bacterial artificial chromosomes are comprised of:
A)components of the bacterial F plasmid and can carry large inserts (200,000 bp).
B)fosmid DNA sequences,enabling the transfer of DNA between bacteria.
C)viral lambda DNA sequences,enabling the transfer of DNA between bacteria.
D)a bacterial chromosome,with some artificial components.
E)recombinant proteins isolated from specific phage viruses.
A)components of the bacterial F plasmid and can carry large inserts (200,000 bp).
B)fosmid DNA sequences,enabling the transfer of DNA between bacteria.
C)viral lambda DNA sequences,enabling the transfer of DNA between bacteria.
D)a bacterial chromosome,with some artificial components.
E)recombinant proteins isolated from specific phage viruses.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
36
Detection of a transgenic plant can be accomplished in a number of different ways,but often the initially transformed cells (in tissue culture)are enriched for the presence of a transgene by:
A)screening groups of cells using the polymerase chain reaction.
B)examining if the phenotype of the cells has been altered.
C)placing the potentially transformed cells in the presence of a drug that inhibits the division of non-transgenic cells.
D)microscopically examining the cells and isolating those that have "extra" DNA.
E)None of the above.Transgenesis cannot be selected for in tissue culture.
A)screening groups of cells using the polymerase chain reaction.
B)examining if the phenotype of the cells has been altered.
C)placing the potentially transformed cells in the presence of a drug that inhibits the division of non-transgenic cells.
D)microscopically examining the cells and isolating those that have "extra" DNA.
E)None of the above.Transgenesis cannot be selected for in tissue culture.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
37
The value of the beta-galactosidase gene in cloning could best be described as the gene:
A)that provides valuable restriction enzyme cut sites.
B)that "reports" if a recombinant product is formed.
C)that confers resistance to antibiotics.
D)that facilitates easier transfer of the DNA into bacterial cells.
E)that encodes a special DNA ligase enzyme.
A)that provides valuable restriction enzyme cut sites.
B)that "reports" if a recombinant product is formed.
C)that confers resistance to antibiotics.
D)that facilitates easier transfer of the DNA into bacterial cells.
E)that encodes a special DNA ligase enzyme.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
38
A cDNA library is produced from ______________ and is valued in the analysis of __________________.
A)chromosomal DNA digested with restriction enzymes;chromosomal organization
B)chromosomal DNA digested with restriction enzymes;the promoter and regulatory elements of a particular gene
C)a DNA copy of mRNA isolated from a group of cells;the protein-encoding open reading frame of a gene (no introns)
D)a DNA copy of mRNA isolated from a group of cells;the promoter and regulatory elements of a particular gene
E)protein sequences isolated from particular tissues;enzyme function encoded by a particular gene
A)chromosomal DNA digested with restriction enzymes;chromosomal organization
B)chromosomal DNA digested with restriction enzymes;the promoter and regulatory elements of a particular gene
C)a DNA copy of mRNA isolated from a group of cells;the protein-encoding open reading frame of a gene (no introns)
D)a DNA copy of mRNA isolated from a group of cells;the promoter and regulatory elements of a particular gene
E)protein sequences isolated from particular tissues;enzyme function encoded by a particular gene

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
39
In Southern and Northern blotting,the probe being used to analyze DNA or RNA identifies the target sequence via:
A)attaching to the gene target via antibody-like protein-to-protein interactions.
B)a DNA ligase-mediated attachment to the target DNA or RNA fragments.
C)annealing between complementary sequences on the probe and the gene target.
D)an enzymatic step that includes crosslinking between similar nucleic acid sequences.
E)amplification during the polymerase chain reaction.
A)attaching to the gene target via antibody-like protein-to-protein interactions.
B)a DNA ligase-mediated attachment to the target DNA or RNA fragments.
C)annealing between complementary sequences on the probe and the gene target.
D)an enzymatic step that includes crosslinking between similar nucleic acid sequences.
E)amplification during the polymerase chain reaction.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
40
When seeking to identify the gene that is responsible for a particular cellular characteristic,scientists will generate cells carrying a recessive mutation that perturbs the characteristic of interest and then try to identify or "clone" the wild-type copy of that gene through functional complementation.This type of complementation could be described as:
A)intensification of the recessive mutant trait via transferred plasmid DNA carrying the gene of interest.
B)reversal (rescue)of the recessive mutant trait via transferred plasmid DNA carrying the gene of interest.
C)identification of two different genes that both regulate the same cellular trait.
D)a biochemical phenomenon where an enzymatic process is eliminated by plasmid DNA expressing a gene of interest.
E)mutation of a gene of interest to complement an opposing biochemical pathway.
A)intensification of the recessive mutant trait via transferred plasmid DNA carrying the gene of interest.
B)reversal (rescue)of the recessive mutant trait via transferred plasmid DNA carrying the gene of interest.
C)identification of two different genes that both regulate the same cellular trait.
D)a biochemical phenomenon where an enzymatic process is eliminated by plasmid DNA expressing a gene of interest.
E)mutation of a gene of interest to complement an opposing biochemical pathway.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
41
You constructed a cDNA library in a lambda vector using mRNAs from a pea plant.You want to identify a rubisco (a protein involved in photosynthesis)clone among the 50,000 clones in the library.You have a rubisco cDNA from tobacco to use as a probe.When hybridizing the probe to the plaques in the library,would you use higher,lower,or the same temperature as for hybridizing the tobacco DNA to a tobacco rubisco clone? Why?

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
42
You purified genomic DNA from tissue isolated from a population of roundworms (C.elegans).You wish to analyze this DNA,so you digest a portion of this DNA with the restriction enzyme EcoRI,and another portion of the DNA is digested with SalI.You make a gel made of agarose,with wells for loading your DNA.You place undigested DNA in well "A," the EcoRI digested DNA in well "B," and the SalI digested DNA in well "C." An electric field is generated by placing an anode (+ electrode)at the base of the gel,and a cathode (- electrode)at the top of the gel near the wells.The DNA migrates towards the anode.
a)Describe the movement of the DNA along the gel.How is an electric field able to generate movement of the DNA fragments?
b)How can nucleic acids be visualized in an agarose gel?
c)How would you expect lane A,B,and C to appear?
d)Can you analyze the size of fragments that contain a particular gene?
a)Describe the movement of the DNA along the gel.How is an electric field able to generate movement of the DNA fragments?
b)How can nucleic acids be visualized in an agarose gel?
c)How would you expect lane A,B,and C to appear?
d)Can you analyze the size of fragments that contain a particular gene?

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
43
A new restriction enzyme is discovered that recognizes an 8-base restriction sequence.About how many fragments of the Wombat genome (approximately 4.2 108 in size)would you expect if you digested it with this enzyme?

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
44
Human insulin can be made purer,and at a lower cost,using recombinant DNA technology.What sort of DNA clone might be ligated into a bacterial expression plasmid to be used in this strategy?

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
45
The restriction enzyme ClaI recognizes the sequence AT CGAT.It cleaves the phosphodiester backbone between the T and the C bases at the arrow.The restriction enzyme TaqI recognizes the sequence T ↓ CGA.It cleaves the phosphodiester backbone between the T and the C bases at the arrow.
a)Indicate the ends of the double-stranded DNA molecule left after cleavage by ClaI.Show the sequence,polarity,and overhang,if any.
b)On average,how many sites would TaqI recognize in a random sequence of 1200 base pairs?
c)If a TaqI-digested DNA end is ligated to a ClaI-digested DNA end,will ClaI cleave the newly ligated DNA? Will TaqI cleave the newly ligated DNA?
a)Indicate the ends of the double-stranded DNA molecule left after cleavage by ClaI.Show the sequence,polarity,and overhang,if any.
b)On average,how many sites would TaqI recognize in a random sequence of 1200 base pairs?
c)If a TaqI-digested DNA end is ligated to a ClaI-digested DNA end,will ClaI cleave the newly ligated DNA? Will TaqI cleave the newly ligated DNA?

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
46
When ligating pieces of DNA into a plasmid,a scientist will often transform the ligation products into competent bacteria to evaluate the success rate of the ligation (i.e.,the number of plasmids that now have extra DNA ligated into their sequence).Define one quick way scientists can assess the success rate of a ligation just by looking at the bacterial colonies.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
47
In a certain community,a mutant allele for Tay-Sachs disease is prevalent.This mutation removes a Hind III restriction site in the gene sequence.A probe is available for this region,with the following homology:
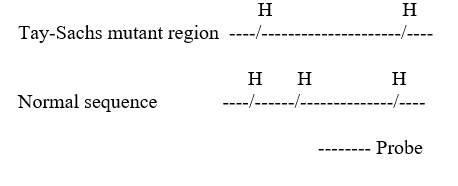
Two couples in this community are expecting children.Each couple has a DNA test to determine if their child will have Tay-Sachs.The results of the Southern blot,probed for the region described above is shown below.
Using these results,how would you counsel the couples?

Two couples in this community are expecting children.Each couple has a DNA test to determine if their child will have Tay-Sachs.The results of the Southern blot,probed for the region described above is shown below.
Using these results,how would you counsel the couples?

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
48
The autoradiogram shown was generated by the dideoxy sequencing method.Define the DNA sequence of the template strand that was used in this sequencing reaction,noting the 5′ and 3′ ends of this template strand. 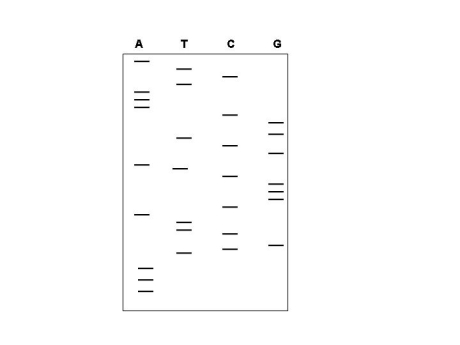


Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
49
A linear molecule of DNA 9 kb in length is isolated and mapped using restriction endonucleases EcoRI and HindIII.The fragments generated by the single and double digests are shown below.The numbers refer to the length of fragments in kilobases.
EcoRI: 7 kb,2 kb
HindIII: 5 kb,4 kb
EcoRI + HindIII: 4 kb,3 kb,2 kb
SalI: 8.5 kb,0.5 kb
EcoRI + SalI: 7 kb,1.5 kb,0.5 kb
Construct a restriction map of this linear fragment of DNA.
EcoRI: 7 kb,2 kb
HindIII: 5 kb,4 kb
EcoRI + HindIII: 4 kb,3 kb,2 kb
SalI: 8.5 kb,0.5 kb
EcoRI + SalI: 7 kb,1.5 kb,0.5 kb
Construct a restriction map of this linear fragment of DNA.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
50
A DNA fragment produced by the restriction enzyme SalI is isolated from a digestion of mouse DNA.You cloned the fragment by first ligating it into a unique Sal I site found in a 3.5 kb cosmid.The ligation into the SalI site regenerates a SalI cut site at each end of the cloned fragment.You decide to determine the restriction map of this fragment.You digest the fragment with different restriction enzymes (below).
Note: The cosmid used in this strategy does not have EcoRI or HindIII sites (but the cloned fragment apparently does).
Note: The cosmid used in this strategy does not have EcoRI or HindIII sites (but the cloned fragment apparently does).

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
51
A certain animal virus contains a circular DNA that has five recognition sites for the restriction enzyme EcoRI.Infected cells make a large quantity of the viral surface protein.A cDNA corresponding to the mRNA for this surface protein has been cloned.How would you use a Southern blot to determine which EcoRI fragment or fragments contain the gene for the viral surface protein?

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
52
Use the following data to generate a restriction map for the circular "pEPP1" plasmid.
PstI: 6.0 kb
EcoRI: 6.0 kb
SalI: 6.0 kb
EcoRI + SalI: 2.0 kb,4.0 kb
EcoRI + PstI: 1.5 kb,4.5 kb
PstI + SalI: 2.5 kb,3.5 kb
PstI + SalI + EcoRI: 1.5 kb,2.0 kb,2.5 kb
PstI: 6.0 kb
EcoRI: 6.0 kb
SalI: 6.0 kb
EcoRI + SalI: 2.0 kb,4.0 kb
EcoRI + PstI: 1.5 kb,4.5 kb
PstI + SalI: 2.5 kb,3.5 kb
PstI + SalI + EcoRI: 1.5 kb,2.0 kb,2.5 kb

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
53
A chimeric plasmid contains vector DNA ligated to a fragment of chromosomal mouse DNA that is suspected of containing the obese gene.As a first step,a restriction map of the DNA clone was constructed.The DNA was digested with two restriction enzymes,BopI and ZapII,with the following results:

a)From these data,answer the following questions.
(1)How large is the entire DNA clone?
(2)Is the clone linear or circular?
(3)Draw a restriction map consistent with these data.Include all BopI and ZapII sites and the physical distances between them.
b)To better identify the location of the mouse chromosomal DNA on this chimeric plasmid,mouse genomic DNA was digested with ZapII,run on a gel,and analyzed using the Southern blot technique.The Southern blot was probed with radiolabeled Zap II DNA fragments from the plasmid DNA.Three lanes each containing the same amount of Zap II-digested chromosomal DNA were each probed with a different fragment from the plasmid.Autoradiograms showed the following: What is the maximum amount of mouse DNA present in the clone? Why didn't the 2.0-kb ZapII fragment hybridize to the mouse DNA?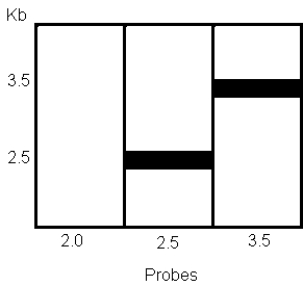

a)From these data,answer the following questions.
(1)How large is the entire DNA clone?
(2)Is the clone linear or circular?
(3)Draw a restriction map consistent with these data.Include all BopI and ZapII sites and the physical distances between them.
b)To better identify the location of the mouse chromosomal DNA on this chimeric plasmid,mouse genomic DNA was digested with ZapII,run on a gel,and analyzed using the Southern blot technique.The Southern blot was probed with radiolabeled Zap II DNA fragments from the plasmid DNA.Three lanes each containing the same amount of Zap II-digested chromosomal DNA were each probed with a different fragment from the plasmid.Autoradiograms showed the following: What is the maximum amount of mouse DNA present in the clone? Why didn't the 2.0-kb ZapII fragment hybridize to the mouse DNA?


Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
54
The drawing below depicts a restriction map of a segment of a human chromosome.The cut sites shown (1 through 6)are known to be polymorphic and can be present or absent depending on the genetic makeup of a specific individual's chromosomes.E identifies EcoRI sites,and P identifies Pst1 sites.The line below the map identifies the region of homology between the DNA and a probe used in Southern blot analyses to characterize this region. 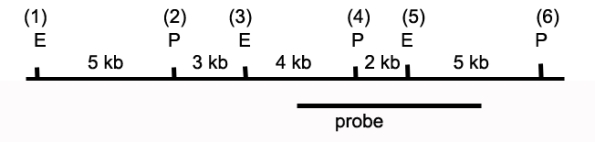 a)Individual 1 has restriction sites 1 through 6 (all)on both copies of this chromosome.If DNA from this individual was digested with Pst I,what are the sizes of the fragments that would be detected via Southern blot using the identified probe?
a)Individual 1 has restriction sites 1 through 6 (all)on both copies of this chromosome.If DNA from this individual was digested with Pst I,what are the sizes of the fragments that would be detected via Southern blot using the identified probe?
b)DNA from Individual 1 is digested with both EcoRI and PstI and then analyzed by Southern blot.What bands are revealed?
c)Individual 2 is homozygous for a polymorphism that eliminates site 3.If DNA from this individual was digested with both PstI and EcoRI,what are the sizes of the fragments that would be detected via Southern blot?
d)Individual 3 has not previously been characterized over this chromosomal locus.The research team digests the DNA with PstI and EcoRI and generates the following banding pattern.What is the likely genotype of this individual?
 a)Individual 1 has restriction sites 1 through 6 (all)on both copies of this chromosome.If DNA from this individual was digested with Pst I,what are the sizes of the fragments that would be detected via Southern blot using the identified probe?
a)Individual 1 has restriction sites 1 through 6 (all)on both copies of this chromosome.If DNA from this individual was digested with Pst I,what are the sizes of the fragments that would be detected via Southern blot using the identified probe?b)DNA from Individual 1 is digested with both EcoRI and PstI and then analyzed by Southern blot.What bands are revealed?
c)Individual 2 is homozygous for a polymorphism that eliminates site 3.If DNA from this individual was digested with both PstI and EcoRI,what are the sizes of the fragments that would be detected via Southern blot?
d)Individual 3 has not previously been characterized over this chromosomal locus.The research team digests the DNA with PstI and EcoRI and generates the following banding pattern.What is the likely genotype of this individual?


Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck
55
A purified DNA preparation of a certain plasmid is digested to completion with BamH1 restriction endonuclease.In separate reactions,the same preparation was digested to completion with EcoRI and with a mixture of EcoRI and BamH1,respectively.The diagram below shows the appearance of the original molecules and the digestion products in the three digestion mixtures,after separation by electrophoresis in an agarose gel.
Lane M-linear DNA markers of the indicated length in kb
Lane 1-sample of original plasmid DNA preparation
Lane 2-sample of the products of BamH1 digestion
Lane 3-sample of the products of EcoRI digestion
Lane 4-sample of the products of digestion of BamH1 + EcoRI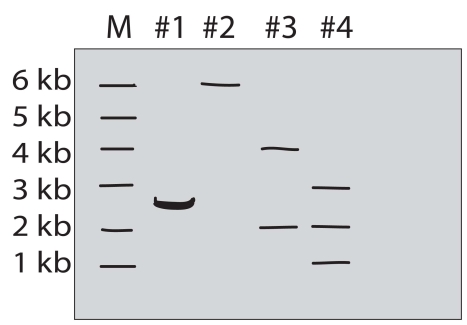
Answer and justify the following:
a)What was the original DNA molecule's length?
b)Was the original plasmid DNA circular or linear?
c)How many restriction sites for EcoRI and BamH1 did the original plasmid have?
d)Draw the original plasmid,using B and E to indicate the location of the BamH1 and EcoRI restriction sites.Indicate the number of kb between sites.
Lane M-linear DNA markers of the indicated length in kb
Lane 1-sample of original plasmid DNA preparation
Lane 2-sample of the products of BamH1 digestion
Lane 3-sample of the products of EcoRI digestion
Lane 4-sample of the products of digestion of BamH1 + EcoRI

Answer and justify the following:
a)What was the original DNA molecule's length?
b)Was the original plasmid DNA circular or linear?
c)How many restriction sites for EcoRI and BamH1 did the original plasmid have?
d)Draw the original plasmid,using B and E to indicate the location of the BamH1 and EcoRI restriction sites.Indicate the number of kb between sites.

Unlock Deck
Unlock for access to all 55 flashcards in this deck.
Unlock Deck
k this deck


