Exam 10: Gene Isolation and Manipulation
Exam 1: The Genetics Revolution22 Questions
Exam 2: Single-Gene Inheritance51 Questions
Exam 3: Independent Assortment of Genes55 Questions
Exam 4: Mapping Eukaryote Chromosomes by Recombination64 Questions
Exam 5: The Genetics of Bacteria and Their Viruses44 Questions
Exam 6: Gene Interaction47 Questions
Exam 7: Dna: Structure and Replication50 Questions
Exam 8: Rna: Transcription and Processing53 Questions
Exam 9: Proteins and Their Synthesis53 Questions
Exam 10: Gene Isolation and Manipulation55 Questions
Exam 11: Regulation of Gene Expression in Bacteria and Their Viruses56 Questions
Exam 12: Regulation of Gene Expression in Eukaryotes46 Questions
Exam 13: The Genetic Control of Development36 Questions
Exam 14: Genomes and Genomics26 Questions
Exam 15: The Dynamic Genome: Transposable Elements32 Questions
Exam 16: Mutation, Repair, and Recombination53 Questions
Exam 17: Large-Scale Chromosomal Changes50 Questions
Exam 18: Population Genetics48 Questions
Exam 19: The Inheritance of Complex Traits36 Questions
Exam 20: Evolution of Genes and Traits54 Questions
Select questions type
A 3-kb supercoiled circular plasmid contains the restriction sites shown below.N and X represent NheI and XbaI sites,respectively;A through F mark six points on the plasmid.A preparation of plasmid DNA was separately digested to completion with XbaI alone,NheI alone,and a mixture of the two enzymes.The products were separated by agarose gel electrophoresis.Draw the predicted appearance of the gel,assuming that you ran the following in the various lanes. 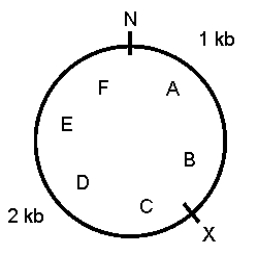 Lane M (size markers)-a mixture of known markers increasing in size from 1 kb to 6 kb,in steps of 1 kb
Lane 1-sample from the original plasmid DNA preparation
Lane 2-products of complete digestion with XbaI alone
Lane 3-products of complete digestion with NheI alone
Lane 4-products of complete digestion with XbaI plus NheI
Lane M (size markers)-a mixture of known markers increasing in size from 1 kb to 6 kb,in steps of 1 kb
Lane 1-sample from the original plasmid DNA preparation
Lane 2-products of complete digestion with XbaI alone
Lane 3-products of complete digestion with NheI alone
Lane 4-products of complete digestion with XbaI plus NheI
Free
(Essay)
4.8/5  (37)
(37)
Correct Answer:
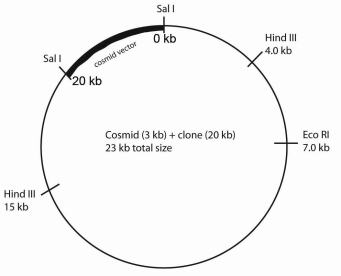
A DNA fragment produced by the restriction enzyme SalI is isolated from a digestion of mouse DNA.You cloned the fragment by first ligating it into a unique Sal I site found in a 3.5 kb cosmid.The ligation into the SalI site regenerates a SalI cut site at each end of the cloned fragment.You decide to determine the restriction map of this fragment.You digest the fragment with different restriction enzymes (below).
Note: The cosmid used in this strategy does not have EcoRI or HindIII sites (but the cloned fragment apparently does).
Restriction enzyme Fragments produced SalI 20,3.5 SalI + EcoRI 7,13,3.5 SalI + HindIII 4,5,11,3.5 SalI + EcoRI + HindIII 3,4,5,8,3.5
Free
(Essay)
4.8/5  (35)
(35)
Correct Answer:
Which of the following statements about PCR is/are FALSE?
Free
(Multiple Choice)
4.8/5  (29)
(29)
Correct Answer:
C
The value of the beta-galactosidase gene in cloning could best be described as the gene:
(Multiple Choice)
4.7/5  (38)
(38)
When seeking to identify the gene that is responsible for a particular cellular characteristic,scientists will generate cells carrying a recessive mutation that perturbs the characteristic of interest and then try to identify or "clone" the wild-type copy of that gene through functional complementation.This type of complementation could be described as:
(Multiple Choice)
4.8/5  (34)
(34)
Assume that a cosmid will carry inserts of about 50 kb and that cosmids are used to clone a 3 Mb (megabase)genome.Assuming you are particularly lucky and have no duplication in your library,what is the smallest number of cosmid clones you would need for a "complete" genomic library?
(Multiple Choice)
4.8/5  (28)
(28)
Thymidine kinase is valuable in the targeting of mouse genes because:
(Multiple Choice)
4.8/5  (38)
(38)
The restriction enzyme ClaI recognizes the sequence AT CGAT.It cleaves the phosphodiester backbone between the T and the C bases at the arrow.The restriction enzyme TaqI recognizes the sequence T ↓ CGA.It cleaves the phosphodiester backbone between the T and the C bases at the arrow.
a)Indicate the ends of the double-stranded DNA molecule left after cleavage by ClaI.Show the sequence,polarity,and overhang,if any.
b)On average,how many sites would TaqI recognize in a random sequence of 1200 base pairs?
c)If a TaqI-digested DNA end is ligated to a ClaI-digested DNA end,will ClaI cleave the newly ligated DNA? Will TaqI cleave the newly ligated DNA?
(Essay)
4.8/5  (27)
(27)
Suppose you cut two different DNAs,
one wih BamH1: 5'-GGATCC-3' 3'-CCTAGG-5' and one with BglII: 5'-A GATCT-3' 3'-TCTAG A-5'
then ligate them together through their compatible sticky ends.Once joined,could you separate these two DNAs again with either restriction enzyme? Why or why not? (Each of these enzymes is cut between the first two nucleotides,at the 5? end of each strand of the palindrome.)
(Essay)
4.8/5  (29)
(29)
Use the following data to generate a restriction map for the circular "pEPP1" plasmid.
PstI: 6.0 kb
EcoRI: 6.0 kb
SalI: 6.0 kb
EcoRI + SalI: 2.0 kb,4.0 kb
EcoRI + PstI: 1.5 kb,4.5 kb
PstI + SalI: 2.5 kb,3.5 kb
PstI + SalI + EcoRI: 1.5 kb,2.0 kb,2.5 kb
(Essay)
4.8/5  (26)
(26)
Transgenic plants can be generated using T-DNA plasmid carrying a gene of interest.To get the DNA into the plant cells,the researchers:
(Multiple Choice)
4.8/5  (35)
(35)
A wild-type Aspergillus strain is transformed with a plasmid carrying a hygromycin resistance allele,and cells are plated on hygromycin.One resistant colony showed an aberrant type of aerial hyphae.When crossed to wild type,the progeny were 1/2 normal hyphae,Hyg sensitive,and 1/2 aberrant hyphae,Hyg-resistant.The probable explanation is that:
(Multiple Choice)
4.9/5  (38)
(38)
You purified genomic DNA from tissue isolated from a population of roundworms (C.elegans).You wish to analyze this DNA,so you digest a portion of this DNA with the restriction enzyme EcoRI,and another portion of the DNA is digested with SalI.You make a gel made of agarose,with wells for loading your DNA.You place undigested DNA in well "A," the EcoRI digested DNA in well "B," and the SalI digested DNA in well "C." An electric field is generated by placing an anode (+ electrode)at the base of the gel,and a cathode (- electrode)at the top of the gel near the wells.The DNA migrates towards the anode.
a)Describe the movement of the DNA along the gel.How is an electric field able to generate movement of the DNA fragments?
b)How can nucleic acids be visualized in an agarose gel?
c)How would you expect lane A,B,and C to appear?
d)Can you analyze the size of fragments that contain a particular gene?
(Essay)
4.8/5  (38)
(38)
A certain animal virus contains a circular DNA that has five recognition sites for the restriction enzyme EcoRI.Infected cells make a large quantity of the viral surface protein.A cDNA corresponding to the mRNA for this surface protein has been cloned.How would you use a Southern blot to determine which EcoRI fragment or fragments contain the gene for the viral surface protein?
(Essay)
4.8/5  (31)
(31)
A new restriction enzyme is discovered that recognizes an 8-base restriction sequence.About how many fragments of the Wombat genome (approximately 4.2 108 in size)would you expect if you digested it with this enzyme?
(Essay)
4.8/5  (40)
(40)
This linear vector is comprised of bacteriophage genome components and can carry cloned DNA up to 15 kb in size.
(Multiple Choice)
4.8/5  (34)
(34)
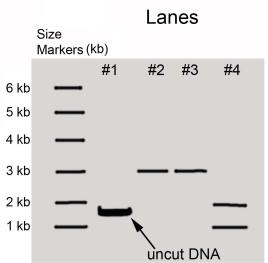
A purified DNA preparation of a certain plasmid is digested to completion with BamH1 restriction endonuclease.In separate reactions,the same preparation was digested to completion with EcoRI and with a mixture of EcoRI and BamH1,respectively.The diagram below shows the appearance of the original molecules and the digestion products in the three digestion mixtures,after separation by electrophoresis in an agarose gel.
Lane M-linear DNA markers of the indicated length in kb
Lane 1-sample of original plasmid DNA preparation
Lane 2-sample of the products of BamH1 digestion
Lane 3-sample of the products of EcoRI digestion
Lane 4-sample of the products of digestion of BamH1 + EcoRI 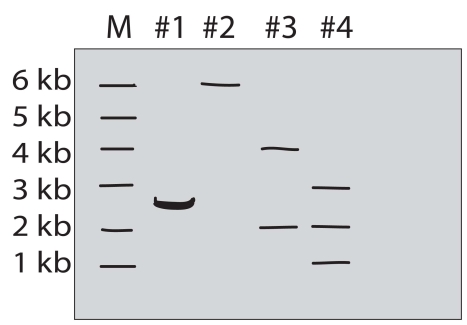 Answer and justify the following:
a)What was the original DNA molecule's length?
b)Was the original plasmid DNA circular or linear?
c)How many restriction sites for EcoRI and BamH1 did the original plasmid have?
d)Draw the original plasmid,using B and E to indicate the location of the BamH1 and EcoRI restriction sites.Indicate the number of kb between sites.
Answer and justify the following:
a)What was the original DNA molecule's length?
b)Was the original plasmid DNA circular or linear?
c)How many restriction sites for EcoRI and BamH1 did the original plasmid have?
d)Draw the original plasmid,using B and E to indicate the location of the BamH1 and EcoRI restriction sites.Indicate the number of kb between sites.
(Essay)
4.8/5  (37)
(37)
Showing 1 - 20 of 55
Filters
- Essay(0)
- Multiple Choice(0)
- Short Answer(0)
- True False(0)
- Matching(0)