Deck 20: The Molecular Revolution: Biotechnology and Beyond
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/70
Play
Full screen (f)
Deck 20: The Molecular Revolution: Biotechnology and Beyond
1
Many identical copies of genes cloned in bacteria are produced as a result of ________.
A) plasmid replication
B) bacterial cell replication
C) transformation
D) plasmid and bacterial cell replication
A) plasmid replication
B) bacterial cell replication
C) transformation
D) plasmid and bacterial cell replication
D
2
In recombinant DNA methods, the term "vector" can refer to ________.
A) the enzyme (endonuclease) that cuts DNA into restriction fragments
B) the "sticky end" of a DNA fragment
C) circular endogenous DNA in mammalian cell
D) a plasmid used to transfer DNA into a living cell
E) a DNA probe used to identify a particular gene
A) the enzyme (endonuclease) that cuts DNA into restriction fragments
B) the "sticky end" of a DNA fragment
C) circular endogenous DNA in mammalian cell
D) a plasmid used to transfer DNA into a living cell
E) a DNA probe used to identify a particular gene
D
3
If a scientist needs to put new DNA into a bacterium, she must ________ the bacterium.
A) transform
B) ligate
C) make a library of the genes of
D) make a cDNA library of
E) make a vector of
A) transform
B) ligate
C) make a library of the genes of
D) make a cDNA library of
E) make a vector of
A
4
Which of the following sequences in double-stranded DNA is most likely to be recognized as a cutting site for a restriction enzyme (endonuclease)?
A) AAGG TTCC
B) AGTC TCAG
C) GGCC CCGG
D) ACCA TGGT
E) AAAA TTTT
A) AAGG TTCC
B) AGTC TCAG
C) GGCC CCGG
D) ACCA TGGT
E) AAAA TTTT

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
5
If mRNAs could be ligated and replicated within plasmids, what enzyme commonly used in recombinant DNA technology would no longer be needed?
A) restriction enzymes
B) DNA polymerase
C) RNA polymerase
D) reverse transcriptase
E) Taq polymerase
A) restriction enzymes
B) DNA polymerase
C) RNA polymerase
D) reverse transcriptase
E) Taq polymerase

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
6
The restriction enzymes BamHI and BclI cut at the points indicated by arrows: 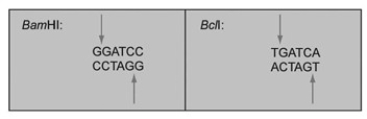 If one sample of DNA was cut with BamHI and another with BclI, and these two samples were mixed and treated with DNA ligase, what would occur?
If one sample of DNA was cut with BamHI and another with BclI, and these two samples were mixed and treated with DNA ligase, what would occur?
A) No DNAs would be ligated (joined together).
B) Only the BamHI-cut DNA would be ligated.
C) Only the BclI-cut DNA would be ligated.
D) Both BamHI-cut and BclI-cut DNAs would be ligated but only to DNA fragments cut with the same enzyme (for example, BamHI fragments would be ligated only to other BamHI fragments).
E) Both BamHI-cut and BclI-cut DNAs would be ligated with no preference for which fragment is ligated to another.
 If one sample of DNA was cut with BamHI and another with BclI, and these two samples were mixed and treated with DNA ligase, what would occur?
If one sample of DNA was cut with BamHI and another with BclI, and these two samples were mixed and treated with DNA ligase, what would occur?A) No DNAs would be ligated (joined together).
B) Only the BamHI-cut DNA would be ligated.
C) Only the BclI-cut DNA would be ligated.
D) Both BamHI-cut and BclI-cut DNAs would be ligated but only to DNA fragments cut with the same enzyme (for example, BamHI fragments would be ligated only to other BamHI fragments).
E) Both BamHI-cut and BclI-cut DNAs would be ligated with no preference for which fragment is ligated to another.

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
7
Which of the following could use reverse transcriptase to transcribe its genome?
A) ssRNA
B) dsRNA
C) ssDNA
D) dsDNA
A) ssRNA
B) dsRNA
C) ssDNA
D) dsDNA

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
8
Which of the following most correctly describes the whole-genome shotgun technique for sequencing a genome?
A) genetic mapping followed immediately by sequencing
B) physical mapping followed immediately by sequencing
C) cloning large genome fragments into very large vectors such as YACs, followed by sequencing
D) cloning fragments from many copies of an entire chromosome, sequencing the fragments, and then ordering the sequences
E) cloning the whole genome directly, from one end to the other
A) genetic mapping followed immediately by sequencing
B) physical mapping followed immediately by sequencing
C) cloning large genome fragments into very large vectors such as YACs, followed by sequencing
D) cloning fragments from many copies of an entire chromosome, sequencing the fragments, and then ordering the sequences
E) cloning the whole genome directly, from one end to the other

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
9
What is a primary difference between polymerase chain reaction (PCR) and traditional cloning procedures such as those used to clone the human growth hormone gene?
A) PCR and traditional cloning make use of different types of bacteria.
B) PCR and traditional cloning make use of different types of vectors.
C) PCR uses plasmid vectors, whereas traditional cloning uses bacteria.
D) PCR eliminates the need for restriction enzymes, vectors, and cells.
E) PCR is more time-consuming, but the purity of the obtained DNA clone is much higher than in traditional cloning.
A) PCR and traditional cloning make use of different types of bacteria.
B) PCR and traditional cloning make use of different types of vectors.
C) PCR uses plasmid vectors, whereas traditional cloning uses bacteria.
D) PCR eliminates the need for restriction enzymes, vectors, and cells.
E) PCR is more time-consuming, but the purity of the obtained DNA clone is much higher than in traditional cloning.

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
10
In a single polymerase chain reaction (PCR) cycle consisting of 15 seconds at 94°C, 30 seconds at 50°C, and 1 minute at 72°C, what is happening in the step run at 50°C?
A) The DNA to be amplified is being denatured.
B) Primers are being denatured.
C) DNA polymerase is extending new DNA from the primers.
D) Primers are annealing to the DNA to be amplified.
E) DNA polymerase is being inactivated.
A) The DNA to be amplified is being denatured.
B) Primers are being denatured.
C) DNA polymerase is extending new DNA from the primers.
D) Primers are annealing to the DNA to be amplified.
E) DNA polymerase is being inactivated.

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
11
Bioinformatics can be used to scan for short sequences that specify known mRNAs, called ________.
A) expressed sequence tags
B) cDNA
C) multigene families
D) proteomes
E) short tandem repeats
A) expressed sequence tags
B) cDNA
C) multigene families
D) proteomes
E) short tandem repeats

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
12
In early forensic applications of DNA fingerprinting, DNA was extracted from crime scene material, digested with restriction enzymes, and then used to prepare a Southern blot. Today, polymerase chain reaction (PCR) is used in the early steps of forensic DNA analysis. Which of the following is an advantage that PCR provides over the former method?
A) PCR produces many more bands for fingerprint analysis, making it a more informative technique.
B) PCR can cut DNA at many more sites than can restriction enzymes.
C) PCR requires much less DNA for analysis.
D) PCR can analyze DNA, proteins, and carbohydrates, whereas restriction enzyme analysis is limited to DNA.
A) PCR produces many more bands for fingerprint analysis, making it a more informative technique.
B) PCR can cut DNA at many more sites than can restriction enzymes.
C) PCR requires much less DNA for analysis.
D) PCR can analyze DNA, proteins, and carbohydrates, whereas restriction enzyme analysis is limited to DNA.

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
13
How do we describe transformation in bacteria?
A) the creation of a strand of DNA from an RNA molecule
B) the creation of a strand of RNA from a DNA molecule
C) the infection of cells by a phage DNA molecule
D) semiconservative replication of DNA
E) insertion of external DNA into a cell
A) the creation of a strand of DNA from an RNA molecule
B) the creation of a strand of RNA from a DNA molecule
C) the infection of cells by a phage DNA molecule
D) semiconservative replication of DNA
E) insertion of external DNA into a cell

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
14
A recent study compared the Homo sapiens genome with that of Neanderthals. The results of the study indicated a mixing of the two genomes at some period in evolutionary history. Additional data consistent with this hypothesis could be the discovery of ________.
A) some Neanderthal sequences not found in living humans
B) a few modern H. sapiens with some Neanderthal sequences
C) duplications of several Neanderthal genes on a Neanderthal chromosome
D) some Neanderthal chromosomes that are shorter than their counterparts in living humans
A) some Neanderthal sequences not found in living humans
B) a few modern H. sapiens with some Neanderthal sequences
C) duplications of several Neanderthal genes on a Neanderthal chromosome
D) some Neanderthal chromosomes that are shorter than their counterparts in living humans

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
15
A paleontologist has recovered a bit of tissue from the 400-year-old preserved skin of an extinct dodo (a bird). To compare a specific region of the DNA from the sample with DNA from living birds, which of the following would be most useful for increasing the amount of dodo DNA available for testing?
A) RFLP analysis
B) polymerase chain reaction (PCR)
C) electroporation
D) gel electrophoresis
E) Southern blotting
A) RFLP analysis
B) polymerase chain reaction (PCR)
C) electroporation
D) gel electrophoresis
E) Southern blotting

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
16
What information is critical to the success of polymerase chain reaction (PCR) itself?
A) The DNA sequence of the ends of the DNA to be amplified must be known.
B) The complete DNA sequence of the DNA to be amplified must be known.
C) The sequence of restriction-enzyme recognition sites in the DNA to be amplified must be known.
D) The sequence of restriction-enzyme recognition sites in the DNA to be amplified and in the plasmid where the amplified DNA fragment will be cloned must be known.
A) The DNA sequence of the ends of the DNA to be amplified must be known.
B) The complete DNA sequence of the DNA to be amplified must be known.
C) The sequence of restriction-enzyme recognition sites in the DNA to be amplified must be known.
D) The sequence of restriction-enzyme recognition sites in the DNA to be amplified and in the plasmid where the amplified DNA fragment will be cloned must be known.

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
17
Use the following information to answer the question(s) below.
In the textbook you read about the amplification and sequencing of 30,000-year-old Neanderthal DNA to learn that it's unlikely Neanderthals and modern humans interbred. Let's take a closer look at this early work with ancient DNA (M. Krings, A. Stone, R.W. Schmitz, H. Krainitzki, M. Stoneking, and S. Pääbo. 1997. Neanderthal DNA sequences and the origin of modern humans. Cell 90:19-30).
Here is part of the lead paragraph in the "Methods" section of the Krings et al. paper:
Protective clothing was worn throughout the sampling procedure. Instruments used were treated with 1 M HCl followed by extensive rinsing in distilled water. After removal, the sample was immediately put into a sterile tube for transport to Munich. All subsequent manipulations of the sample, and experimental procedures prior to cycling of PCR reactions, were carried out in laboratories solely dedicated to the analysis of archaeological specimens, where protective clothing, separate equipment and reagents, UV irradiation, and other measures to... .
A concluding paragraph of the Krings et al. paper states:
It must be emphasized that the above conclusions are based on a single individual sequence; the retrieval and analysis of mtDNA sequences from additional Neanderthal specimens is obviously desirable. If this proves possible, then the potential exists to address several questions concerning Neanderthals that hitherto could be studied exclusively by morphological and archaeological approaches. For example, the genetic relationship between Neanderthal populations in Europe and in western Asia could be explored, as could the demographic history of Neanderthal populations.
Refer to the paragraphs on the Krings et al. paper. How could the "demographic history of Neanderthal populations" be explored if mitochondrial DNA from other Neanderthal samples could be PCR amplified?
A) These Neanderthal sequences could be compared to those of modern humans to learn how modern humans evolved to walk on two legs (bipedalism).
B) These Neanderthal sequences could be compared to those of modern humans; the more they differ, the more distant the time of divergence between the Neanderthal and modern human lineages.
C) These Neanderthal sequences could be compared to each other to see how much they differ; the more they differ, the greater the divergence between these populations.
D) In the attempt to amplify other Neanderthal DNAs, those that amplify must be from more modern populations and those that don't are from more ancient populations.
In the textbook you read about the amplification and sequencing of 30,000-year-old Neanderthal DNA to learn that it's unlikely Neanderthals and modern humans interbred. Let's take a closer look at this early work with ancient DNA (M. Krings, A. Stone, R.W. Schmitz, H. Krainitzki, M. Stoneking, and S. Pääbo. 1997. Neanderthal DNA sequences and the origin of modern humans. Cell 90:19-30).
Here is part of the lead paragraph in the "Methods" section of the Krings et al. paper:
Protective clothing was worn throughout the sampling procedure. Instruments used were treated with 1 M HCl followed by extensive rinsing in distilled water. After removal, the sample was immediately put into a sterile tube for transport to Munich. All subsequent manipulations of the sample, and experimental procedures prior to cycling of PCR reactions, were carried out in laboratories solely dedicated to the analysis of archaeological specimens, where protective clothing, separate equipment and reagents, UV irradiation, and other measures to... .
A concluding paragraph of the Krings et al. paper states:
It must be emphasized that the above conclusions are based on a single individual sequence; the retrieval and analysis of mtDNA sequences from additional Neanderthal specimens is obviously desirable. If this proves possible, then the potential exists to address several questions concerning Neanderthals that hitherto could be studied exclusively by morphological and archaeological approaches. For example, the genetic relationship between Neanderthal populations in Europe and in western Asia could be explored, as could the demographic history of Neanderthal populations.
Refer to the paragraphs on the Krings et al. paper. How could the "demographic history of Neanderthal populations" be explored if mitochondrial DNA from other Neanderthal samples could be PCR amplified?
A) These Neanderthal sequences could be compared to those of modern humans to learn how modern humans evolved to walk on two legs (bipedalism).
B) These Neanderthal sequences could be compared to those of modern humans; the more they differ, the more distant the time of divergence between the Neanderthal and modern human lineages.
C) These Neanderthal sequences could be compared to each other to see how much they differ; the more they differ, the greater the divergence between these populations.
D) In the attempt to amplify other Neanderthal DNAs, those that amplify must be from more modern populations and those that don't are from more ancient populations.

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
18
What characteristic of short tandem repeats (STRs) DNA makes it useful for DNA fingerprinting?
A) The number of repeats varies widely from person to person or animal to animal.
B) The sequence of DNA that is repeated varies significantly from individual to individual.
C) The sequence variation is acted upon differently by natural selection in different environments.
D) Every racial and ethnic group has inherited different short tandem repeats.
A) The number of repeats varies widely from person to person or animal to animal.
B) The sequence of DNA that is repeated varies significantly from individual to individual.
C) The sequence variation is acted upon differently by natural selection in different environments.
D) Every racial and ethnic group has inherited different short tandem repeats.

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
19
Use the following information to answer the question(s) below.
In the textbook you read about the amplification and sequencing of 30,000-year-old Neanderthal DNA to learn that it's unlikely Neanderthals and modern humans interbred. Let's take a closer look at this early work with ancient DNA (M. Krings, A. Stone, R.W. Schmitz, H. Krainitzki, M. Stoneking, and S. Pääbo. 1997. Neanderthal DNA sequences and the origin of modern humans. Cell 90:19-30).
Here is part of the lead paragraph in the "Methods" section of the Krings et al. paper:
Protective clothing was worn throughout the sampling procedure. Instruments used were treated with 1 M HCl followed by extensive rinsing in distilled water. After removal, the sample was immediately put into a sterile tube for transport to Munich. All subsequent manipulations of the sample, and experimental procedures prior to cycling of PCR reactions, were carried out in laboratories solely dedicated to the analysis of archaeological specimens, where protective clothing, separate equipment and reagents, UV irradiation, and other measures to... .
A concluding paragraph of the Krings et al. paper states:
It must be emphasized that the above conclusions are based on a single individual sequence; the retrieval and analysis of mtDNA sequences from additional Neanderthal specimens is obviously desirable. If this proves possible, then the potential exists to address several questions concerning Neanderthals that hitherto could be studied exclusively by morphological and archaeological approaches. For example, the genetic relationship between Neanderthal populations in Europe and in western Asia could be explored, as could the demographic history of Neanderthal populations.
Refer to the paragraph on the Krings et al. paper. Why did the researchers wear protective clothing and note so emphatically that they did so?
A) They needed to be certain there were no ancient pathogens on the sample that modern humans hadn't been exposed to.
B) They needed to be sure not to harm the precious sample with oils from skin or moisture from their breath.
C) They needed to minimize the chance of introducing their own mitochondrial DNA to the sample.
D) They needed to follow careful, standardized laboratory procedures required by law and document their compliance.
In the textbook you read about the amplification and sequencing of 30,000-year-old Neanderthal DNA to learn that it's unlikely Neanderthals and modern humans interbred. Let's take a closer look at this early work with ancient DNA (M. Krings, A. Stone, R.W. Schmitz, H. Krainitzki, M. Stoneking, and S. Pääbo. 1997. Neanderthal DNA sequences and the origin of modern humans. Cell 90:19-30).
Here is part of the lead paragraph in the "Methods" section of the Krings et al. paper:
Protective clothing was worn throughout the sampling procedure. Instruments used were treated with 1 M HCl followed by extensive rinsing in distilled water. After removal, the sample was immediately put into a sterile tube for transport to Munich. All subsequent manipulations of the sample, and experimental procedures prior to cycling of PCR reactions, were carried out in laboratories solely dedicated to the analysis of archaeological specimens, where protective clothing, separate equipment and reagents, UV irradiation, and other measures to... .
A concluding paragraph of the Krings et al. paper states:
It must be emphasized that the above conclusions are based on a single individual sequence; the retrieval and analysis of mtDNA sequences from additional Neanderthal specimens is obviously desirable. If this proves possible, then the potential exists to address several questions concerning Neanderthals that hitherto could be studied exclusively by morphological and archaeological approaches. For example, the genetic relationship between Neanderthal populations in Europe and in western Asia could be explored, as could the demographic history of Neanderthal populations.
Refer to the paragraph on the Krings et al. paper. Why did the researchers wear protective clothing and note so emphatically that they did so?
A) They needed to be certain there were no ancient pathogens on the sample that modern humans hadn't been exposed to.
B) They needed to be sure not to harm the precious sample with oils from skin or moisture from their breath.
C) They needed to minimize the chance of introducing their own mitochondrial DNA to the sample.
D) They needed to follow careful, standardized laboratory procedures required by law and document their compliance.

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
20
Which of the following is in the correct order for one cycle of polymerase chain reaction (PCR)?
A) Denature DNA; add fresh enzyme; anneal primers; add dNTPs; extend primers.
B) Add fresh enzyme; denature DNA; anneal primers; add dNTPs; extend primers.
C) Anneal primers; denature DNA; extend primers.
D) Extend primers; anneal primers; denature DNA.
E) Denature DNA; anneal primers; extend primers.
A) Denature DNA; add fresh enzyme; anneal primers; add dNTPs; extend primers.
B) Add fresh enzyme; denature DNA; anneal primers; add dNTPs; extend primers.
C) Anneal primers; denature DNA; extend primers.
D) Extend primers; anneal primers; denature DNA.
E) Denature DNA; anneal primers; extend primers.

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
21
Metagenomics aims to learn about the diversity of organisms, particularly microbes that inhabit natural environments. J. Craig Venter, a key figure in the race to obtain the human genome sequence, and his colleagues pioneered this approach in a study that analyzed DNA obtained from microbes collected from the Sargasso Sea, an intensively studied, nutrient-impoverished part of the Caribbean lying to the southeast of Bermuda. (C. J. Venter, K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkock, Y.-H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74.)
Refer to the paragraph on the Venter et al. paper and the accompanying table. How do you think expected genes were placed into the categories shown in the table?
A) by searching for open reading frames (ORFs), especially those with translation start and stop sites
B) by searching for matches, especially of the predicted gene products, with previously identified gene products of known function
C) by expressing the cloned genes in E. coli and then running biochemical analyses of the products
D) by searching for similarity between ORFs and those in existing databases
Refer to the paragraph on the Venter et al. paper and the accompanying table. How do you think expected genes were placed into the categories shown in the table?
A) by searching for open reading frames (ORFs), especially those with translation start and stop sites
B) by searching for matches, especially of the predicted gene products, with previously identified gene products of known function
C) by expressing the cloned genes in E. coli and then running biochemical analyses of the products
D) by searching for similarity between ORFs and those in existing databases

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
22
Bioinformatics includes ________.
I) using computer programs to align DNA sequences
II) creating recombinant DNA from separate species
III) developing computer-based tools for genome analysis
IV) using mathematical tools to make sense of biological systems
A) I and II
B) II and III
C) II and IV
D) I, III, and IV
I) using computer programs to align DNA sequences
II) creating recombinant DNA from separate species
III) developing computer-based tools for genome analysis
IV) using mathematical tools to make sense of biological systems
A) I and II
B) II and III
C) II and IV
D) I, III, and IV

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
23
The best type of scientist to organize a large amount of genome sequence data would be someone who specializes in ________.
A) proteomics
B) functional genomics
C) evolutionary genomics
D) bioinformatics
A) proteomics
B) functional genomics
C) evolutionary genomics
D) bioinformatics

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
24
Metagenomics aims to learn about the diversity of organisms, particularly microbes that inhabit natural environments. J. Craig Venter, a key figure in the race to obtain the human genome sequence, and his colleagues pioneered this approach in a study that analyzed DNA obtained from microbes collected from the Sargasso Sea, an intensively studied, nutrient-impoverished part of the Caribbean lying to the southeast of Bermuda. (C. J. Venter, K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkock, Y.-H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74.)
Refer to the paragraph on the Venter et al. paper and the accompanying table. As with many new endeavors, concerns have been raised by some that genomics will fundamentally change the way biological research is practiced. Examine the full citation for the Venter et al. paper and consider the way this work was done. Now think of some seminal works in the history of biology, for example, Darwin's On the Origin of Species and Watson and Crick's publication on DNA structure. What do you think the concern might be that has been voiced about genomics?
A) Genomics will offer deep insights too rapidly for the biological community to comprehend.
B) Genomics is so based in technology that it cannot possibly reveal important knowledge of biology.
C) Genomics makes biology a "big-team science" that will squeeze out innovative individuals or small groups of scientists who traditionally have provided our greatest insights.
D) Genomics is really computer science, not biology, and therefore offers little for understanding life.
Refer to the paragraph on the Venter et al. paper and the accompanying table. As with many new endeavors, concerns have been raised by some that genomics will fundamentally change the way biological research is practiced. Examine the full citation for the Venter et al. paper and consider the way this work was done. Now think of some seminal works in the history of biology, for example, Darwin's On the Origin of Species and Watson and Crick's publication on DNA structure. What do you think the concern might be that has been voiced about genomics?
A) Genomics will offer deep insights too rapidly for the biological community to comprehend.
B) Genomics is so based in technology that it cannot possibly reveal important knowledge of biology.
C) Genomics makes biology a "big-team science" that will squeeze out innovative individuals or small groups of scientists who traditionally have provided our greatest insights.
D) Genomics is really computer science, not biology, and therefore offers little for understanding life.

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
25
Metagenomics aims to learn about the diversity of organisms, particularly microbes that inhabit natural environments. J. Craig Venter, a key figure in the race to obtain the human genome sequence, and his colleagues pioneered this approach in a study that analyzed DNA obtained from microbes collected from the Sargasso Sea, an intensively studied, nutrient-impoverished part of the Caribbean lying to the southeast of Bermuda. (C. J. Venter, K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkock, Y.-H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74.)
Refer to the paragraph on the Venter et al. paper and the accompanying table. What practical applications for human society may arise first from this or similar metagenomic studies?
A) New genes with useful properties may be identified and ultimately put to use.
B) New microbial species could be isolated from seawater.
C) An increased knowledge of the diversity of marine ecosystems could lead to investigations of diversity in other ecosystems.
D) Species discovered in the Sargasso Sea may also be found to inhabit less-remote waters.
Refer to the paragraph on the Venter et al. paper and the accompanying table. What practical applications for human society may arise first from this or similar metagenomic studies?
A) New genes with useful properties may be identified and ultimately put to use.
B) New microbial species could be isolated from seawater.
C) An increased knowledge of the diversity of marine ecosystems could lead to investigations of diversity in other ecosystems.
D) Species discovered in the Sargasso Sea may also be found to inhabit less-remote waters.

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
26
The comparison between the number of human genes and those of other animal species has led to many conclusions, including that ________.
A) the density of the human genome is far higher than in most other animals
B) the number of proteins expressed by the human genome is far more than the number of its genes
C) most human DNA consists of genes for protein, tRNA, rRNA, and miRNA
D) the genomes of most other organisms are significantly smaller than the human genome
A) the density of the human genome is far higher than in most other animals
B) the number of proteins expressed by the human genome is far more than the number of its genes
C) most human DNA consists of genes for protein, tRNA, rRNA, and miRNA
D) the genomes of most other organisms are significantly smaller than the human genome

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
27
Sequencing eukaryotic genomes is more difficult than sequencing genomes of bacteria or archaea because of the ________.
A) large size of eukaryotic proteins
B) hard-to-find proteins
C) high proportion of G-C base pairs in eukaryotic DNA
D) large size of eukaryotic genomes and the large amount of eukaryotic repetitive DNA
A) large size of eukaryotic proteins
B) hard-to-find proteins
C) high proportion of G-C base pairs in eukaryotic DNA
D) large size of eukaryotic genomes and the large amount of eukaryotic repetitive DNA

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
28
After finding a new medicinal plant, a pharmaceutical company decides to determine if the plant has genes similar to those of other known medicinal plants. To do this, the company annotates the genome of the new plant to ________.
A) determine what proteins are produced
B) determine what mRNA transcripts are produced
C) identify genes and determine their functions
D) identify the location of mRNA within the plant cells
A) determine what proteins are produced
B) determine what mRNA transcripts are produced
C) identify genes and determine their functions
D) identify the location of mRNA within the plant cells

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
29
The bulk of the sequence data in whole-genome sequencing directly comes from ________.
A) relatively large (~160 kb) sequences cloned into bacterial artificial chromosomes (BACs)
B) relatively small (~1000 base-pair) sequences cloned into plasmids
C) whole chromosomes obtained without cloning
D) whole chromosomes cloned into plasmids
A) relatively large (~160 kb) sequences cloned into bacterial artificial chromosomes (BACs)
B) relatively small (~1000 base-pair) sequences cloned into plasmids
C) whole chromosomes obtained without cloning
D) whole chromosomes cloned into plasmids

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
30
Unequal crossing over during prophase I can result in one sister chromosome with a deletion and another with a duplication. A mutated form of hemoglobin, so-called hemoglobin Lepore, exists in the human population. Hemoglobin Lepore has a deleted series of amino acids. If this mutated form was caused by unequal crossing over, what would be an expected consequence?
A) If it is still maintained in the human population, hemoglobin Lepore must be selected for in evolution.
B) There should also be persons whose hemoglobin contains two copies of the series of amino acids that is deleted in hemoglobin Lepore.
C) Each of the genes in the hemoglobin gene family must show the same deletion.
D) The deleted gene must have undergone exon shuffling.
E) The deleted region must be located in a different area of the individual's genome.
A) If it is still maintained in the human population, hemoglobin Lepore must be selected for in evolution.
B) There should also be persons whose hemoglobin contains two copies of the series of amino acids that is deleted in hemoglobin Lepore.
C) Each of the genes in the hemoglobin gene family must show the same deletion.
D) The deleted gene must have undergone exon shuffling.
E) The deleted region must be located in a different area of the individual's genome.

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
31
A laboratory might use dideoxyribonucleotides to ________.
A) separate DNA fragments
B) clone the breakpoints of cut DNA
C) produce cDNA from mRNA
D) sequence a DNA fragment
E) visualize DNA expression
A) separate DNA fragments
B) clone the breakpoints of cut DNA
C) produce cDNA from mRNA
D) sequence a DNA fragment
E) visualize DNA expression

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
32
Biologists now routinely test for homology between genes in different species. If genes are determined to be homologous, it means that they are related ________.
A) by descent from a common ancestor
B) because of convergent evolution
C) by chance mutations
D) in function but not structure
A) by descent from a common ancestor
B) because of convergent evolution
C) by chance mutations
D) in function but not structure

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
33
An early step in shotgun sequencing is to ________.
A) break genomic DNA at random sites
B) map the position of cloned DNA fragments
C) randomly select DNA primers and hybridize these to random positions of chromosomes in preparation for sequencing
D) separate molecules of DNA according to size in an electric field
A) break genomic DNA at random sites
B) map the position of cloned DNA fragments
C) randomly select DNA primers and hybridize these to random positions of chromosomes in preparation for sequencing
D) separate molecules of DNA according to size in an electric field

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
34
Metagenomics aims to learn about the diversity of organisms, particularly microbes that inhabit natural environments. J. Craig Venter, a key figure in the race to obtain the human genome sequence, and his colleagues pioneered this approach in a study that analyzed DNA obtained from microbes collected from the Sargasso Sea, an intensively studied, nutrient-impoverished part of the Caribbean lying to the southeast of Bermuda. (C. J. Venter, K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkock, Y.-H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74.)
Refer to the paragraph on the Venter et al. paper and the accompanying table. Because DNAs for sequencing were chosen at random with no way of knowing which organism they came from, how could genes be identified?
A) by searching for open reading frames (ORFs), especially those with translation start and stop sites
B) by searching for similarity between predicted ORFs and those in existing databases
C) by searching for exons bounded by consensus donor and acceptor splice sites (those that mark exon/intron boundaries)
D) by searching for ORFs, particularly those with translation start and stop sites, as well as for similarity between predicted ORFs and those in existing databases
Refer to the paragraph on the Venter et al. paper and the accompanying table. Because DNAs for sequencing were chosen at random with no way of knowing which organism they came from, how could genes be identified?
A) by searching for open reading frames (ORFs), especially those with translation start and stop sites
B) by searching for similarity between predicted ORFs and those in existing databases
C) by searching for exons bounded by consensus donor and acceptor splice sites (those that mark exon/intron boundaries)
D) by searching for ORFs, particularly those with translation start and stop sites, as well as for similarity between predicted ORFs and those in existing databases

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
35
A multigene family is composed of ________.
A) multiple genes whose products must be coordinately expressed
B) genes whose sequences are very similar and that probably arose by duplication
C) many tandem repeats such as those found in centromeres and telomeres
D) a gene whose exons can be spliced in a number of different ways
E) a highly conserved gene found in a number of different species
A) multiple genes whose products must be coordinately expressed
B) genes whose sequences are very similar and that probably arose by duplication
C) many tandem repeats such as those found in centromeres and telomeres
D) a gene whose exons can be spliced in a number of different ways
E) a highly conserved gene found in a number of different species

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
36
What is metagenomics?
A) genomics as applied to a species that most typifies the average phenotype of its genus
B) the sequencing of one or two representative genes from several species
C) the sequencing of only the most highly conserved genes in a lineage
D) sequencing DNA from a group of species from the same ecosystem
E) genomics as applied to an entire phylum
A) genomics as applied to a species that most typifies the average phenotype of its genus
B) the sequencing of one or two representative genes from several species
C) the sequencing of only the most highly conserved genes in a lineage
D) sequencing DNA from a group of species from the same ecosystem
E) genomics as applied to an entire phylum

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
37
Sequencing an entire genome, such as that of C. elegans, a nematode, is most important because ________.
A) it allows researchers to use the sequence to build a "better" nematode, which is resistant to disease
B) it allows research on a group of organisms we do not usually care much about
C) the nematode is a good animal model for trying out cures for viral illness
D) a sequence that is found to have a particular function in the nematode is likely to have a closely related function in vertebrates
E) a sequence that is found to have no introns in the nematode genome is likely to have acquired the introns from higher organisms
A) it allows researchers to use the sequence to build a "better" nematode, which is resistant to disease
B) it allows research on a group of organisms we do not usually care much about
C) the nematode is a good animal model for trying out cures for viral illness
D) a sequence that is found to have a particular function in the nematode is likely to have a closely related function in vertebrates
E) a sequence that is found to have no introns in the nematode genome is likely to have acquired the introns from higher organisms

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
38
It is more difficult to identify eukaryotic genes than prokaryotic genes because in eukaryotes ________.
A) the proteins are larger than in prokaryotes
B) the coding portions of genes are shorter than in prokaryotes
C) there are no start codons
D) there are introns
A) the proteins are larger than in prokaryotes
B) the coding portions of genes are shorter than in prokaryotes
C) there are no start codons
D) there are introns

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
39
When comparing the genomes of a bacterial parasite versus a free-living, nonparasitic bacterium, one expects to find that ________.
A) the proportion of G-C base pairs will be significantly higher in the parasite
B) there is no homology between the free-living and parasitic bacteria
C) there will be a higher proportion of genes of unknown function in the parasite
D) the parasite will have acquired a smaller proportion of its genes through lateral gene transfer
E) the parasite will have a smaller genome
A) the proportion of G-C base pairs will be significantly higher in the parasite
B) there is no homology between the free-living and parasitic bacteria
C) there will be a higher proportion of genes of unknown function in the parasite
D) the parasite will have acquired a smaller proportion of its genes through lateral gene transfer
E) the parasite will have a smaller genome

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
40
If the sequence of a cDNA has matches with DNA sequences in the genome, then this genomic DNA is likely to ________.
A) code for a protein
B) code for a ribozyme
C) code for an rRNA
D) be part of an intron
E) be a regulatory sequence
A) code for a protein
B) code for a ribozyme
C) code for an rRNA
D) be part of an intron
E) be a regulatory sequence

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
41
In nature, the major mechanism of introducing new genes into eukaryotic genomes is ________.
A) duplication followed by evolutionary divergence
B) unequal crossing over at microsatellite repeats
C) pseudogene creation
D) pseudogene restoration
A) duplication followed by evolutionary divergence
B) unequal crossing over at microsatellite repeats
C) pseudogene creation
D) pseudogene restoration

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
42
Chronic myelogenous leukemia (CML) results from a translocation between human chromosomes 9 and 22. The resulting chromosome 22 is significantly shorter than usual, and it is known as a Philadelphia (Ph') chromosome. The junction at the site of the translocation causes overexpression of a receptor tyrosine kinase. A new drug (Gleevec or imatinib) has been found to inhibit the disease if the patient is treated early. Which of the following would be a reasonably efficient technique for confirming the diagnosis of CML?
A) searching for the number of telomeric sequences on chromosome 22
B) looking for a Ph' chromosome in a peripheral blood smear
C) enzyme assay for receptor tyrosine kinase activity
D) fluorescent labeling to determine the chromosomal location of all chromosome 22 fragments
E) identification of the disease phenotype in review of the patient's records
A) searching for the number of telomeric sequences on chromosome 22
B) looking for a Ph' chromosome in a peripheral blood smear
C) enzyme assay for receptor tyrosine kinase activity
D) fluorescent labeling to determine the chromosomal location of all chromosome 22 fragments
E) identification of the disease phenotype in review of the patient's records

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
43
Because they both produce a reverse transcriptase, long interspersed nuclear elements (LINEs) as transposable elements may be related to ________.
A) plasmids
B) retroviruses
C) poliovirus
D) parasitic bacteria
E) lower plants
A) plasmids
B) retroviruses
C) poliovirus
D) parasitic bacteria
E) lower plants

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
44
What is the difference between a linkage map and a physical map?
A) Markers are spaced by recombination frequency on a linkage map and by number of base pairs on a physical map.
B) The ATCG order and sequence must be determined for a linkage map but not for a physical map.
C) A linkage map shows how each gene is linked to every other gene, but a physical map does not.
D) Distances must be calculable in units such as nanometers on a physical map but not on a linkage map.
E) There is no difference between the two except in the type of pictorial representation.
A) Markers are spaced by recombination frequency on a linkage map and by number of base pairs on a physical map.
B) The ATCG order and sequence must be determined for a linkage map but not for a physical map.
C) A linkage map shows how each gene is linked to every other gene, but a physical map does not.
D) Distances must be calculable in units such as nanometers on a physical map but not on a linkage map.
E) There is no difference between the two except in the type of pictorial representation.

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
45
If alternative splicing did NOT occur, then ________.
A) the human genome would likely contain many more genes
B) the E. coli genome would contain many fewer genes
C) there would be little correlation between the complexity of organisms and genome size
D) there would be many fewer genes devoted to metabolism in Arabidopsis and yeast
A) the human genome would likely contain many more genes
B) the E. coli genome would contain many fewer genes
C) there would be little correlation between the complexity of organisms and genome size
D) there would be many fewer genes devoted to metabolism in Arabidopsis and yeast

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
46
Which of the following is one of the technical reasons why gene therapy is problematic?
A) Most cells with an engineered gene do not produce gene product.
B) Most cells with engineered genes overwhelm other cells in a tissue.
C) Cells with transferred genes are unlikely to replicate.
D) Transferred genes may not have appropriately controlled activity.
E) mRNA from transferred genes cannot be translated.
A) Most cells with an engineered gene do not produce gene product.
B) Most cells with engineered genes overwhelm other cells in a tissue.
C) Cells with transferred genes are unlikely to replicate.
D) Transferred genes may not have appropriately controlled activity.
E) mRNA from transferred genes cannot be translated.

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
47
Which viruses have single-stranded RNA that acts as a template for DNA synthesis?
A) lytic phages
B) proviruses
C) viroids
D) bacteriophages
E) retroviruses
A) lytic phages
B) proviruses
C) viroids
D) bacteriophages
E) retroviruses

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
48
Use the following information to answer the question(s) below.
Six students have taken samples of their own cheek cells, purified the DNA, and used an enzyme (endonuclease) known to cut at zero, one, or two sites in a particular gene of interest.
Their next two steps, in order, should be ________.
A) use of a fluorescent probe for the gene sequence, then electrophoresis
B) electrophoresis of the fragments, followed by autoradiography
C) electrophoresis of the fragments, followed by the use of a probe
D) use of a ligase that will anneal the pieces, followed by Southern blotting
E) use of reverse transcriptase to make cDNA, followed by electrophoresis
Six students have taken samples of their own cheek cells, purified the DNA, and used an enzyme (endonuclease) known to cut at zero, one, or two sites in a particular gene of interest.
Their next two steps, in order, should be ________.
A) use of a fluorescent probe for the gene sequence, then electrophoresis
B) electrophoresis of the fragments, followed by autoradiography
C) electrophoresis of the fragments, followed by the use of a probe
D) use of a ligase that will anneal the pieces, followed by Southern blotting
E) use of reverse transcriptase to make cDNA, followed by electrophoresis

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
49
Use the following information to answer the question(s) below.
Six students have taken samples of their own cheek cells, purified the DNA, and used an enzyme (endonuclease) known to cut at zero, one, or two sites in a particular gene of interest.
They might be conducting such an experiment to ________.
A) find the location of a particular gene in the human genome
B) prepare to isolate the chromosome on which there is a particular gene of interest
C) find which of the students has which alleles
D) measure the expression of one or more genes in their genomes
E) determine if any of them carries an extra sex chromosome
Six students have taken samples of their own cheek cells, purified the DNA, and used an enzyme (endonuclease) known to cut at zero, one, or two sites in a particular gene of interest.
They might be conducting such an experiment to ________.
A) find the location of a particular gene in the human genome
B) prepare to isolate the chromosome on which there is a particular gene of interest
C) find which of the students has which alleles
D) measure the expression of one or more genes in their genomes
E) determine if any of them carries an extra sex chromosome

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
50
Imagine that you are searching for the gene associated with nail-patella syndrome, a dominant genetic disorder that causes developmental abnormalities. In a large pedigree you discover an association between nail-patella syndrome and a genetic marker that occurs in two different alleles, A and B. Fifteen individuals within this pedigree have nail-patella syndrome and are A/B heterozygotes for the marker. Thirty individuals within this pedigree don't suffer from nail-patella syndrome and are homozygous for the A marker allele. One individual within this pedigree has nail-patella syndrome and is also homozygous for the A marker allele. The most likely explanation for this exceptional individual is that ________.
A) a new mutation converted the disease-causing allele to the wild-type form
B) a new mutation converted the B allele of the marker to the A form
C) the exceptional nail-patella individual is haploid
D) recombination occurred between the nail-patella gene and marker locus in one of the parents of the exceptional individual
A) a new mutation converted the disease-causing allele to the wild-type form
B) a new mutation converted the B allele of the marker to the A form
C) the exceptional nail-patella individual is haploid
D) recombination occurred between the nail-patella gene and marker locus in one of the parents of the exceptional individual

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
51
In large-scale, genome-wide association studies in humans we look for ________.
A) lengthy sequences that might be shared by most members of a population
B) SNPs where one allele is found more often in persons with a particular disorder than in healthy controls
C) SNPs where one allele is found in families with a particular introns sequence
D) SNPs where one allele is found in two or more adjacent genes
E) large inversions that displace the centromere
A) lengthy sequences that might be shared by most members of a population
B) SNPs where one allele is found more often in persons with a particular disorder than in healthy controls
C) SNPs where one allele is found in families with a particular introns sequence
D) SNPs where one allele is found in two or more adjacent genes
E) large inversions that displace the centromere

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
52
Analysis of the data obtained shows that two students each have two fragments, two students each have three fragments, and two students each have one only. What does this demonstrate?
A) Each pair of students has a different gene for this function.
B) The two students who have two fragments have one restriction site in this region.
C) The two students who have two fragments have two restriction sites within this gene.
D) The students with three fragments are said to have "fragile sites."
E) Each of these students is heterozygous for this gene.
A) Each pair of students has a different gene for this function.
B) The two students who have two fragments have one restriction site in this region.
C) The two students who have two fragments have two restriction sites within this gene.
D) The students with three fragments are said to have "fragile sites."
E) Each of these students is heterozygous for this gene.

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
53
The mechanism for the spread of long interspersed nuclear elements (LINEs) involves some unique features, including ________.
A) translation in the cytoplasm of an mRNA that was synthesized in the nucleus
B) the nuclear localization of RNA polymerase
C) the transport of mRNA from the cytoplasm to the nucleus
D) the use of an enzyme that cuts DNA
E) the use of RNA polymerase to transcribe an mRNA
A) translation in the cytoplasm of an mRNA that was synthesized in the nucleus
B) the nuclear localization of RNA polymerase
C) the transport of mRNA from the cytoplasm to the nucleus
D) the use of an enzyme that cuts DNA
E) the use of RNA polymerase to transcribe an mRNA

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
54
Imagine that you compare two DNA sequences found in the same location on homologous chromosomes. On one of the homologs, the sequence is AACTACGA. On the other homolog, the sequence is AACTTCGA. Within a population, you discover that each of these sequences is common. These sequences ________.
A) contain an SNP that may be useful for genetic mapping
B) identify a protein-coding region of a gene
C) cause disease
D) protect against disease
E) do none of the listed actions
A) contain an SNP that may be useful for genetic mapping
B) identify a protein-coding region of a gene
C) cause disease
D) protect against disease
E) do none of the listed actions

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
55
Name two examples of lateral gene transfer to eukaryotes that involve cellular organelles.
A) transfer of bacterial genes that were predecessors to mitochondria and chloroplasts
B) transfer of the bacterial mitochondrial apparatus to form nuclei and ribosomes
C) transfer of bacterial chloroplasts to form nuclei and ribosomes
D) transfer of bacterial genes to form nuclei and the mitotic spindle apparatus
E) transfer of the bacterial spindle apparatus to form ribosomes and transfer RNAs
A) transfer of bacterial genes that were predecessors to mitochondria and chloroplasts
B) transfer of the bacterial mitochondrial apparatus to form nuclei and ribosomes
C) transfer of bacterial chloroplasts to form nuclei and ribosomes
D) transfer of bacterial genes to form nuclei and the mitotic spindle apparatus
E) transfer of the bacterial spindle apparatus to form ribosomes and transfer RNAs

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
56
Transgenic mice are useful to human researchers because they ________.
A) can be valuable animal models of human disease
B) are essential for mapping human genes
C) are now used in place of bacteria for cloning human genes
D) were instrumental in pinpointing the location of the huntingtin gene
A) can be valuable animal models of human disease
B) are essential for mapping human genes
C) are now used in place of bacteria for cloning human genes
D) were instrumental in pinpointing the location of the huntingtin gene

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
57
Viruses use the host's machinery to make copies of themselves. However, some human viruses require a type of replication that humans do not normally have. For example, humans normally do not have the ability to convert RNA into DNA. How can these types of viruses infect humans, when human cells cannot perform a particular role that the virus requires?
A) The virus causes mutations in the human cells, resulting in the formation of new enzymes that are capable of performing these roles.
B) The viral genome codes for specialized enzymes not in the host.
C) The virus infects only those cells and species that can perform all the replication roles necessary.
D) Viruses can stay in a quiescent state until the host cell evolves this ability.
A) The virus causes mutations in the human cells, resulting in the formation of new enzymes that are capable of performing these roles.
B) The viral genome codes for specialized enzymes not in the host.
C) The virus infects only those cells and species that can perform all the replication roles necessary.
D) Viruses can stay in a quiescent state until the host cell evolves this ability.

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
58
If you are a human geneticist looking for an ideal population in which to map disease genes, which of the following characteristics would help the mapping study?
A) small family sizes with well-known genealogies
B) a population where all members have the disease
C) large family sizes with little known genealogies
D) large family sizes with well-known genealogies
A) small family sizes with well-known genealogies
B) a population where all members have the disease
C) large family sizes with little known genealogies
D) large family sizes with well-known genealogies

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
59
What is the function of reverse transcriptase in retroviruses?
A) It hydrolyzes the host cell's DNA.
B) It uses viral RNA as a template for DNA synthesis.
C) It converts host cell RNA into viral DNA.
D) It translates viral RNA into proteins.
E) It uses viral RNA as a template for making complementary RNA strands.
A) It hydrolyzes the host cell's DNA.
B) It uses viral RNA as a template for DNA synthesis.
C) It converts host cell RNA into viral DNA.
D) It translates viral RNA into proteins.
E) It uses viral RNA as a template for making complementary RNA strands.

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
60
A virus consisting of a single strand of RNA, which is transcribed into complementary DNA, is a ________.
A) reverse transcriptase
B) protease
C) retrovirus
D) RNA replicase virus
E) nonenveloped virus
A) reverse transcriptase
B) protease
C) retrovirus
D) RNA replicase virus
E) nonenveloped virus

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
61
Proteomics is defined as the ________.
A) linkage of each gene to a particular protein
B) study of the full protein set encoded by a genome
C) totality of the functional possibilities of a single protein
D) study of how amino acids are ordered in a protein
E) study of how a single gene activates many proteins
A) linkage of each gene to a particular protein
B) study of the full protein set encoded by a genome
C) totality of the functional possibilities of a single protein
D) study of how amino acids are ordered in a protein
E) study of how a single gene activates many proteins

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
62
Which type of disorder is most difficult to correct by gene therapy?
A) a dominant disorder
B) an incompletely dominant disorder
C) a recessive disorder
D) Disorders showing all of these forms of dominance present equal challenges.
A) a dominant disorder
B) an incompletely dominant disorder
C) a recessive disorder
D) Disorders showing all of these forms of dominance present equal challenges.

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
63
For applications in gene therapy, what is the most favorable characteristic of retroviruses?
A) Retroviruses have an RNA genome.
B) Retroviruses possess reverse transcriptase.
C) DNA copies of retroviral genomes become integrated into the genome of the infected cell.
D) Retroviruses mutate often.
A) Retroviruses have an RNA genome.
B) Retroviruses possess reverse transcriptase.
C) DNA copies of retroviral genomes become integrated into the genome of the infected cell.
D) Retroviruses mutate often.

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
64
The genes needed for β-carotene synthesis are not normally expressed in endosperm. In the creation of golden rice, how did researchers ensure that these genes were expressed in rice endosperm?
A) The genes were injected only in the endosperm of rice grains.
B) Agrobacterium cells were added only to rice grains, not to any other part of the plant.
C) The protein-coding sequence of each gene was linked to a regulatory region of DNA that directed transcription in endosperm.
D) The wild-type genes were mutated and new mutant alleles that are expressed only in endosperm were selected.
A) The genes were injected only in the endosperm of rice grains.
B) Agrobacterium cells were added only to rice grains, not to any other part of the plant.
C) The protein-coding sequence of each gene was linked to a regulatory region of DNA that directed transcription in endosperm.
D) The wild-type genes were mutated and new mutant alleles that are expressed only in endosperm were selected.

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
65
Gene therapy requires ________.
A) knowledge and availability of the normal allele of the defective gene
B) the ability to introduce the normal allele into the patient
C) the ability to express the introduced gene at the correct level, time, and tissue site within the patient
D) knowledge and availability of the normal allele of the defective gene and the ability to introduce the allele into the patient
E) knowledge and availability of the normal allele of the defective gene, an ability to introduce the normal allele into the patient, and an ability to express the introduced gene at the correct level, and time, and tissue site within the patient
A) knowledge and availability of the normal allele of the defective gene
B) the ability to introduce the normal allele into the patient
C) the ability to express the introduced gene at the correct level, time, and tissue site within the patient
D) knowledge and availability of the normal allele of the defective gene and the ability to introduce the allele into the patient
E) knowledge and availability of the normal allele of the defective gene, an ability to introduce the normal allele into the patient, and an ability to express the introduced gene at the correct level, and time, and tissue site within the patient

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
66
DNA microarrays have made a huge impact on genomic studies because they ________.
A) can be used to eliminate the function of any gene in the genome
B) can be used to introduce entire genomes into bacterial cells
C) allow the expression of many or even all of the genes in the genome to be compared at once
D) allow physical maps of the genome to be assembled in a very short time
E) dramatically enhance the efficiency of restriction enzymes (endonucleases)
A) can be used to eliminate the function of any gene in the genome
B) can be used to introduce entire genomes into bacterial cells
C) allow the expression of many or even all of the genes in the genome to be compared at once
D) allow physical maps of the genome to be assembled in a very short time
E) dramatically enhance the efficiency of restriction enzymes (endonucleases)

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
67
Which of the following questions is asked in functional genomics research?
A) What is the number of Drosophila genes?
B) How does the G-C content of human DNA vary across the genome?
C) What is the pattern of gene expression during mouse development?
D) How many introns exist in the human CFTR gene?
E) How closely related are the visual pigment genes of mouse and human?
A) What is the number of Drosophila genes?
B) How does the G-C content of human DNA vary across the genome?
C) What is the pattern of gene expression during mouse development?
D) How many introns exist in the human CFTR gene?
E) How closely related are the visual pigment genes of mouse and human?

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
68
What can proteomics reveal that genomics cannot?
A) the number of genes characteristic of a species
B) the patterns of alternative splicing
C) the set of proteins present within a cell or tissue type
D) the levels of mRNAs present in a particular cell type
E) the movement of transposable elements within the genome
A) the number of genes characteristic of a species
B) the patterns of alternative splicing
C) the set of proteins present within a cell or tissue type
D) the levels of mRNAs present in a particular cell type
E) the movement of transposable elements within the genome

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
69
One example of an innovation from an unexpected source comes from studies of tumor-like plant growths. What information did the study of plant tumors provide that was critical for plant genetic engineering?
A) understanding the role of plant hormones in growth promotion
B) knowledge of plant tumor suppressor and proto-oncogenes
C) learning that metastasis is critical for tumor progression
D) learning the biosynthetic pathway to β-carotene
E) discovery of a plasmid that could be modified to introduce genes into plants
A) understanding the role of plant hormones in growth promotion
B) knowledge of plant tumor suppressor and proto-oncogenes
C) learning that metastasis is critical for tumor progression
D) learning the biosynthetic pathway to β-carotene
E) discovery of a plasmid that could be modified to introduce genes into plants

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck
70
The areas that have received the greatest attention in genetically modified plants are ________.
A) improved product quality and increased pest and herbicide resistance
B) improved product quality, more rapid growth rate through plant hormone production, and decreased herbicide resistance
C) more rapid growth rate through plant hormone production, pharmaceutical production, and increased antibiotic resistance
D) pharmaceutical production, production of useful compounds not normally found in nature (xenobiotics), and the ability to remove or neutralize toxic compounds from the environment (bioremediation)
A) improved product quality and increased pest and herbicide resistance
B) improved product quality, more rapid growth rate through plant hormone production, and decreased herbicide resistance
C) more rapid growth rate through plant hormone production, pharmaceutical production, and increased antibiotic resistance
D) pharmaceutical production, production of useful compounds not normally found in nature (xenobiotics), and the ability to remove or neutralize toxic compounds from the environment (bioremediation)

Unlock Deck
Unlock for access to all 70 flashcards in this deck.
Unlock Deck
k this deck


