Deck 15: Dna Technologies
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/91
Play
Full screen (f)
Deck 15: Dna Technologies
1
How are the sticky ends obtained?
A)by using a different restriction enzyme to cut both DNA fragment and the plasmid
B)by using the same restriction enzyme to cut both DNA fragment and the plasmid
C)by using a restriction enzyme to cut DNA fragment
D)by using a restriction enzyme to cut the plasmid
A)by using a different restriction enzyme to cut both DNA fragment and the plasmid
B)by using the same restriction enzyme to cut both DNA fragment and the plasmid
C)by using a restriction enzyme to cut DNA fragment
D)by using a restriction enzyme to cut the plasmid
B
2
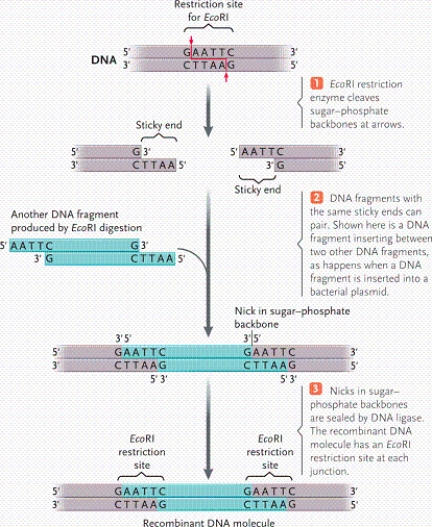
According to the table, which enzyme was used to digest a DNA fragment that results in the following sequence? AATTC---------G
G---------CTTAA
A)EcoRI
B)ClaI
C)BamHI
D)HindIII
A
3
Which of the following describes DNA ligase?
A)the enzyme that seals together DNA fragments generated by restriction enzyme digestion to produce an original DNA molecule
B)the enzyme that seals together DNA fragments generated by restriction enzyme digestion to produce a recombinant DNA molecule
C)the enzyme that seals together RNA fragments generated by restriction enzyme digestion to produce a recombinant DNA molecule
D)the enzyme that seals together RNA fragments generated by restriction enzyme digestion to produce a recombinant RNA molecule
A)the enzyme that seals together DNA fragments generated by restriction enzyme digestion to produce an original DNA molecule
B)the enzyme that seals together DNA fragments generated by restriction enzyme digestion to produce a recombinant DNA molecule
C)the enzyme that seals together RNA fragments generated by restriction enzyme digestion to produce a recombinant DNA molecule
D)the enzyme that seals together RNA fragments generated by restriction enzyme digestion to produce a recombinant RNA molecule
B
4
What is the natural function of restriction endonucleases?
A)regulating bacterial gene expression
B)DNA recombination in vivo
C)defending against viruses that infect bacteria
D)DNA manipulation in vitro
A)regulating bacterial gene expression
B)DNA recombination in vivo
C)defending against viruses that infect bacteria
D)DNA manipulation in vitro

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
5

According to the table, which enzymes should be used to generate a double-stranded sequence that looks like the one below? AGCTT---------G
A---------CCTAG
A)ClaI and PvuI
B)BamHI and ClaI
C)EcoRI and HindIII
D)HindIII and BamHI

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
6
How many sources of DNA sequences are commonly used in biotechnology?
A)1
B)2
C)3
D)4
A)1
B)2
C)3
D)4

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
7
If you were to utilize "gene machine," which of the following would you be using?
A)a machine that can create DNA sequences of choice by automated chemical synthesis in the laboratory using a RNA synthesizer responding to computer input
B)a machine that can create DNA sequences of choice by manual chemical synthesis in the laboratory using a DNA synthesizer responding to computer input
C)a machine that can create DNA sequences of choice by automated chemical synthesis in the laboratory using a DNA synthesizer responding to computer input
D)a machine that can create RNA sequences of choice by automated chemical synthesis in the laboratory using a RNA synthesizer responding to computer input
A)a machine that can create DNA sequences of choice by automated chemical synthesis in the laboratory using a RNA synthesizer responding to computer input
B)a machine that can create DNA sequences of choice by manual chemical synthesis in the laboratory using a DNA synthesizer responding to computer input
C)a machine that can create DNA sequences of choice by automated chemical synthesis in the laboratory using a DNA synthesizer responding to computer input
D)a machine that can create RNA sequences of choice by automated chemical synthesis in the laboratory using a RNA synthesizer responding to computer input

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
8

According to the table, which enzymes were used to digest a DNA fragment that results in the following sequence? GATCC---------AT
G---------TAGC
A)BamHI and ClaI
B)HindIII and BamHI
C)EcoRI and HindIII
D)ClaI and PvuI

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
9
Some restriction endonucleases cut the DNA in such a way that short, single-stranded regions are created. What are these regions called?
A)blunt ends
B)tacky ends
C)hydrogen-bonding ends
D)sticky ends
A)blunt ends
B)tacky ends
C)hydrogen-bonding ends
D)sticky ends

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
10
Which of the following refer to nonrecombinant plasmids?
A)resealed cloning vectors with no DNA fragment inserted
B)plasmids with DNA fragments inserted into the cloning vector
C)plaids with no DNA fragment inserted
D)resealed cloning vectors with DNA fragments inserted into the cloning vector
A)resealed cloning vectors with no DNA fragment inserted
B)plasmids with DNA fragments inserted into the cloning vector
C)plaids with no DNA fragment inserted
D)resealed cloning vectors with DNA fragments inserted into the cloning vector

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
11
Why is there X-gal used in the cloning medium?
A)It distinguishes between bacteria that have been transformed with recombinant plasmids and nonrecombinant plasmids by black-white screening.
B)It distinguishes between bacteria that have been transformed with recombinant plasmids and nonrecombinant plasmids by blue-white screening.
C)It distinguishes between plasmids that have been transformed and those that have not by blue-white screening.
D)It distinguishes between plasmids that have been transformed and those that have not by black-white screening.
A)It distinguishes between bacteria that have been transformed with recombinant plasmids and nonrecombinant plasmids by black-white screening.
B)It distinguishes between bacteria that have been transformed with recombinant plasmids and nonrecombinant plasmids by blue-white screening.
C)It distinguishes between plasmids that have been transformed and those that have not by blue-white screening.
D)It distinguishes between plasmids that have been transformed and those that have not by black-white screening.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
12
How long is the DNA sequence that is recognized by restriction endonucleases?
A)generally 1 to 2 nucleotides
B)generally 4 to 8 nucleotides
C)generally 10 to 15 nucleotides
D)generally 24 to 32 nucleotides
A)generally 1 to 2 nucleotides
B)generally 4 to 8 nucleotides
C)generally 10 to 15 nucleotides
D)generally 24 to 32 nucleotides

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
13
Where is cloned DNA in bacteria usually found?
A)on a virus
B)on the flagellum
C)on a plasmid
D)on the bacterial chromosome
A)on a virus
B)on the flagellum
C)on a plasmid
D)on the bacterial chromosome

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
14
In what way are basic and applied research studies of a cloned gene different?
A)In basic research, a researcher wants to study the gene's function, including how its expression is regulated, and the nature of the gene's product, while in applied research, the interest is in using cloned genes for private application.
B)In basic research, a researcher does not want to study the gene's function, including how its expression is regulated, and the nature of the gene's product, while in applied research, the interest is in using cloned genes for medical application.
C)In basic research, a researcher does not learn about the gene's structure, including its DNA sequence and sequences that regulate its expression, while in applied research, the interest is in the structure and function of a gene.
D)In basic research, a researcher learns about the gene's structure, including its DNA sequence and sequences that regulate its expression, while in applied research, the interest is not in the structure and function of a gene.
A)In basic research, a researcher wants to study the gene's function, including how its expression is regulated, and the nature of the gene's product, while in applied research, the interest is in using cloned genes for private application.
B)In basic research, a researcher does not want to study the gene's function, including how its expression is regulated, and the nature of the gene's product, while in applied research, the interest is in using cloned genes for medical application.
C)In basic research, a researcher does not learn about the gene's structure, including its DNA sequence and sequences that regulate its expression, while in applied research, the interest is in the structure and function of a gene.
D)In basic research, a researcher learns about the gene's structure, including its DNA sequence and sequences that regulate its expression, while in applied research, the interest is not in the structure and function of a gene.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
15
Why would researchers want to have both genomic libraries and cDNA libraries from a particular organism?
A)because a cDNA library helps us understand how retroviruses work, but a genomic library does not
B)because a genomic library is generally too big to be helpful, while a cDNA library, being smaller, is much easier to study
C)because a genomic library will be the same in every cell, while a cDNA library tells researchers which genes are being expressed in individual cell types
D)because a cDNA library is a lot easier to clone than a genomic library
A)because a cDNA library helps us understand how retroviruses work, but a genomic library does not
B)because a genomic library is generally too big to be helpful, while a cDNA library, being smaller, is much easier to study
C)because a genomic library will be the same in every cell, while a cDNA library tells researchers which genes are being expressed in individual cell types
D)because a cDNA library is a lot easier to clone than a genomic library

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
16
Which of the following is needed in order to amplify the DNA for gene X and NOT for gene Y?
A)a heat-stable DNA polymerase
B)restriction endonucleases that cut gene X and not gene Y
C)plasmid vectors that have multiple antibiotic resistance genes
D)some prior knowledge of the sequence of gene X such that appropriate primers can be designed
A)a heat-stable DNA polymerase
B)restriction endonucleases that cut gene X and not gene Y
C)plasmid vectors that have multiple antibiotic resistance genes
D)some prior knowledge of the sequence of gene X such that appropriate primers can be designed

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
17
The list below contains steps for cloning a gene of interest. They are in random order, and some may be incorrect. Which sequence places the correct stems in the correct order?
1)Transform the DNA into E. coli.
2)Ligate cut genomic DNA fragments and cut plasmid DNA together using DNA ligase.
3)Digest the plasmid cloning vector with the same restriction enzyme.
4)Isolate genomic DNA and digest that DNA with a restriction enzyme.
5)Spread bacterial cells on medium containing lactose and ampicillin and incubate to allow colonies to grow.
6)Spread the bacterial cells on growth medium containing ampicillin and X-gal and incubate to allow colonies to grow.
A)4, 3, 2, 1, 5
B)4, 2, 3, 1, 5
C)4, 2, 3, 1, 6
D)4, 3, 2, 1, 6
1)Transform the DNA into E. coli.
2)Ligate cut genomic DNA fragments and cut plasmid DNA together using DNA ligase.
3)Digest the plasmid cloning vector with the same restriction enzyme.
4)Isolate genomic DNA and digest that DNA with a restriction enzyme.
5)Spread bacterial cells on medium containing lactose and ampicillin and incubate to allow colonies to grow.
6)Spread the bacterial cells on growth medium containing ampicillin and X-gal and incubate to allow colonies to grow.
A)4, 3, 2, 1, 5
B)4, 2, 3, 1, 5
C)4, 2, 3, 1, 6
D)4, 3, 2, 1, 6

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
18
Why is it important to have an antibiotic resistance gene in the cloning plasmid?
A)It makes the bacteria resistant to the antibiotic.
B)It provides a way to sort bacteria that have the gene of interest from the bacteria that have just an "empty" cloning plasmid.
C)It provides a way for researchers to sort the bacteria that have the cloning plasmid from the bacteria that do not have the cloning plasmid.
D)It aids scientists in developing new antibiotic treatments.
A)It makes the bacteria resistant to the antibiotic.
B)It provides a way to sort bacteria that have the gene of interest from the bacteria that have just an "empty" cloning plasmid.
C)It provides a way for researchers to sort the bacteria that have the cloning plasmid from the bacteria that do not have the cloning plasmid.
D)It aids scientists in developing new antibiotic treatments.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
19
What do restriction endonucleases do?
a.break glycosidic bonds
b.break ester bonds
c.break phosphodiester bonds
d.break peptide bonds
a.break glycosidic bonds
b.break ester bonds
c.break phosphodiester bonds
d.break peptide bonds

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
20
Which of the following describes biotechnology?
A)any technique applied to dead organisms to make or modify products or processes for a specific purpose.
B)any technique applied to non-biological systems or dead organisms to make or modify products or processes for a specific purpose
C)any technique applied to biological systems or living organisms to make or modify products or processes for a specific purpose
D)any technique applied to non-biological systems to make or modify products or processes for a specific purpose
A)any technique applied to dead organisms to make or modify products or processes for a specific purpose.
B)any technique applied to non-biological systems or dead organisms to make or modify products or processes for a specific purpose
C)any technique applied to biological systems or living organisms to make or modify products or processes for a specific purpose
D)any technique applied to non-biological systems to make or modify products or processes for a specific purpose

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
21
What has genetic engineering been used for?
A)to create mice with altered genomes so we can model certain human diseases
B)to produce a vaccine against hoof-and-mouth disease
C)to create plants that are more resistant to certain pests or herbicides
D)to induce bacteria to mass-produce human insulin
A)to create mice with altered genomes so we can model certain human diseases
B)to produce a vaccine against hoof-and-mouth disease
C)to create plants that are more resistant to certain pests or herbicides
D)to induce bacteria to mass-produce human insulin

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
22
Which sequence presents the three stages of a PCR in the correct order?
A)annealing extension
extension  denaturation
denaturation
B)extension denaturation
denaturation  annealing
annealing
C)denaturation annealing
annealing  extension
extension
D)denaturation extension
extension  annealing
annealing
A)annealing
 extension
extension  denaturation
denaturationB)extension
 denaturation
denaturation  annealing
annealingC)denaturation
 annealing
annealing  extension
extensionD)denaturation
 extension
extension  annealing
annealing
Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
23
What are restriction fragment length polymorphisms used for?
A)to prepare DNA for further sequence analysis
B)to treat sickle cell anemia
C)to test the effectiveness of different restriction enzymes on a sequence of DNA
D)to compare DNA from two individuals
A)to prepare DNA for further sequence analysis
B)to treat sickle cell anemia
C)to test the effectiveness of different restriction enzymes on a sequence of DNA
D)to compare DNA from two individuals

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
24
Which of the following is characteristic of cDNA?
A)a double stranded DNA copy of a single-stranded mRNA molecule
B)a single stranded DNA copy of a single-stranded mRNA molecule
C)a single stranded mRNA copy of a double-stranded mRNA molecule
D)a single stranded mRNA copy of a single-stranded mRNA molecule
A)a double stranded DNA copy of a single-stranded mRNA molecule
B)a single stranded DNA copy of a single-stranded mRNA molecule
C)a single stranded mRNA copy of a double-stranded mRNA molecule
D)a single stranded mRNA copy of a single-stranded mRNA molecule

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
25
Suppose that multiple fragments from a genome have been cloned into bacteria. How can researchers identify the bacteria that contain the particular sequence they are interested in?
A)by sequencing all the bacterial DNA
B)by DNA hybridization, using a labelled probe
C)by observing that the bacteria will produce white colonies on X-gal
D)by isolating the DNA of interest before transformation
A)by sequencing all the bacterial DNA
B)by DNA hybridization, using a labelled probe
C)by observing that the bacteria will produce white colonies on X-gal
D)by isolating the DNA of interest before transformation

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
26
Suppose that you generate cDNA libraries for the same cell line under different conditions. Also suppose that when you compare the libraries, you find that some cDNAs in one library are missing from the other library. What is the best explanation for this result?
A)One of the cDNA libraries is really a genomic library.
B)There was an error in generating one of the cDNA libraries, and some of the sequences were not cloned properly.
C)Even the same cell type will change gene expression as conditions change. The difference in cDNAs tells which genes are expressed in each set of conditions.
D)The probe used in the Southern blot of the libraries must have been poorly constructed.
A)One of the cDNA libraries is really a genomic library.
B)There was an error in generating one of the cDNA libraries, and some of the sequences were not cloned properly.
C)Even the same cell type will change gene expression as conditions change. The difference in cDNAs tells which genes are expressed in each set of conditions.
D)The probe used in the Southern blot of the libraries must have been poorly constructed.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
27
to identify samples with specific DNA sequences
Which of the following can be determined by DNA fingerprinting?
A)the sequence of human genome
B)the genome of human fingers
C)the difference between two individuals of the same species
D)the species which a sample came from
Which of the following can be determined by DNA fingerprinting?
A)the sequence of human genome
B)the genome of human fingers
C)the difference between two individuals of the same species
D)the species which a sample came from

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
28
Which of the following best describes the purpose of creating transgenic mice?
A)to study genetic changes in context
B)to produce proteins
C)to clone mice
D)to create antibiotic-resistant mice
A)to study genetic changes in context
B)to produce proteins
C)to clone mice
D)to create antibiotic-resistant mice

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
29
Which of the following is like a set of clones that collectively contains a copy of every DNA sequence in a genome?
A)restriction enzyme library
B)cloning vector
C)genomic DNA library
D)cDNA library
A)restriction enzyme library
B)cloning vector
C)genomic DNA library
D)cDNA library

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
30
Sticky ends are ends on DNA fragments that are complementary to the DNA ends on the open plasmid.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
31
Suppose that you have cloned a genomic library and know that the bacteria have foreign DNA fragments inserted into the cloning vector. Also suppose that you need to identify the specific bacteria that have the gene coding for protein X. How can these bacteria be located?
A)by restriction endonuclease treatment of DNA from some of the bacterial colonies
B)by DNA hybridization of the different colonies
C)by agarose gel electrophoresis of DNA isolated from the bacterial colonies
D)by PCR of the same bacterial mixture that was spread on agarose plates
A)by restriction endonuclease treatment of DNA from some of the bacterial colonies
B)by DNA hybridization of the different colonies
C)by agarose gel electrophoresis of DNA isolated from the bacterial colonies
D)by PCR of the same bacterial mixture that was spread on agarose plates

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
32
Which of the following distinguishes DNA cloning from gene cloning?
A)DNA cloning is a method for producing a few copies of a piece of DNA. Gene cloning is DNA cloning that involves a gene.
B)DNA cloning is a method for producing a piece of DNA. Gene cloning is DNA cloning that involves a gene.
C)Gene cloning is a method for producing many copies of a piece of DNA. DNA cloning is cloning that involves a gene.
D)DNA cloning is a method for producing many copies of a piece of DNA. Gene cloning is DNA cloning that involves a gene.
A)DNA cloning is a method for producing a few copies of a piece of DNA. Gene cloning is DNA cloning that involves a gene.
B)DNA cloning is a method for producing a piece of DNA. Gene cloning is DNA cloning that involves a gene.
C)Gene cloning is a method for producing many copies of a piece of DNA. DNA cloning is cloning that involves a gene.
D)DNA cloning is a method for producing many copies of a piece of DNA. Gene cloning is DNA cloning that involves a gene.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
33
What is a benefit of introducing new genes into a plant?
A)The plant's growth rate reduces.
B)The plant's genes mutate faster.
C)The plant's nutritional value to primary consumers improves.
D)The plant's reliance on photosynthesis reduces.
A)The plant's growth rate reduces.
B)The plant's genes mutate faster.
C)The plant's nutritional value to primary consumers improves.
D)The plant's reliance on photosynthesis reduces.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
34
How is the Ti plasmid used to create transgenic plants?
A)The Ti plasmid inserts the foreign gene directly into the plant's nuclear DNA.
B)Once the Rhizobium radiobacter forms a tumour in the plant, the tumour cells can take up the foreign DNA and incorporate it into their chromosomes.
C)The Ti plasmid allows all of the plant cells to host the bacteria Rhizobium radiobacter and thus express the foreign gene that was inserted into the bacterial chromosome.
D)The Ti plasmid inserts the foreign DNA into the DNA in the chloroplast.
A)The Ti plasmid inserts the foreign gene directly into the plant's nuclear DNA.
B)Once the Rhizobium radiobacter forms a tumour in the plant, the tumour cells can take up the foreign DNA and incorporate it into their chromosomes.
C)The Ti plasmid allows all of the plant cells to host the bacteria Rhizobium radiobacter and thus express the foreign gene that was inserted into the bacterial chromosome.
D)The Ti plasmid inserts the foreign DNA into the DNA in the chloroplast.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
35
In which stage is the foreign gene introduced into the developing embryo during the creation of transgenic animals?
A)at the blastocyst stage, via a transgenic cell
B)in cell culture, prior to fertilization
C)at the sperm stage, via direct microinjection of the new DNA
D)at the egg stage, via direct microinjection of the new DNA
A)at the blastocyst stage, via a transgenic cell
B)in cell culture, prior to fertilization
C)at the sperm stage, via direct microinjection of the new DNA
D)at the egg stage, via direct microinjection of the new DNA

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
36
What are the three essential pieces of DNA required for PCR?
A)one primer and a template
B)two primers and a template
C)one primer and unknown DNA
D)two primers and unknown DNA
A)one primer and a template
B)two primers and a template
C)one primer and unknown DNA
D)two primers and unknown DNA

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
37
What is the natural function of the T DNA contained within the Ti plasmid?
A)to increase diversity
B)to induce tumours
C)to allow for recombination
D)to transfer genes
A)to increase diversity
B)to induce tumours
C)to allow for recombination
D)to transfer genes

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
38
Which approach would be most helpful in determining whether the gene for a particular protein is the same length in frogs, humans, and trees?
A)use cDNA library construction and agarose gel electrophoresis
B)use genomic library construction and PCR
C)use DNA cloning and DNA hybridization
D)use PCR and agarose gel electrophoresis
A)use cDNA library construction and agarose gel electrophoresis
B)use genomic library construction and PCR
C)use DNA cloning and DNA hybridization
D)use PCR and agarose gel electrophoresis

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
39
What is the main difference between germ-line gene therapy and somatic gene therapy?
A)Only germ-line gene therapy will have results limited to the current generation.
B)Only somatic gene therapy can result in the modified genes being passed to the next generation.
C)Only germ-line gene therapy results in altered gametes.
D)Only the germ-line cells are altered in somatic gene therapy.
A)Only germ-line gene therapy will have results limited to the current generation.
B)Only somatic gene therapy can result in the modified genes being passed to the next generation.
C)Only germ-line gene therapy results in altered gametes.
D)Only the germ-line cells are altered in somatic gene therapy.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
40
What drawback have researchers noted in the cloning of domestic animals?
A)The cloned animals often suffer from birth defects.
B)The cloned animals do not resemble the parent.
C)The cloned animals do not reach adulthood.
D)The cloned animals are not viable.
A)The cloned animals often suffer from birth defects.
B)The cloned animals do not resemble the parent.
C)The cloned animals do not reach adulthood.
D)The cloned animals are not viable.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
41
Gene therapy is a safe, effective, and commercially available treatment for numerous genetic disorders.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
42
Match each term with its definition.
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
functional genomics
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
functional genomics

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
43
Some genetically modified tobacco plants can express the light-producing chemicals present in fireflies.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
44
Different methodologies can often be used to obtain the same information about a DNA sequence.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
45
Explain the ethical concerns with germ-line gene therapy in humans.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
46
When using embryonic germ-line cells to create transgenic mice, the offspring of the genetically modified animal will have all transgenic cells in their bodies.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
47
Match each term with its definition.
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
restriction endonucleases
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
restriction endonucleases

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
48
Match each term with its definition.
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
DNA hybridization
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
DNA hybridization

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
49
DNA fingerprinting is limited to human studies.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
50
Gene therapy has been successful in curing some individuals of autoimmune disorder and sickle cell disease.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
51
Make an argument for which molecular biology technique discussed in this chapter is the single most important technique for forensic scientists and justify your answer with the benefits of this method compared to the others that are available to you.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
52
Genetically modified crops have been banned in Canada.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
53
Define the purpose of cloning a gene into a plasmid cloning vector.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
54
Match each term with its definition.
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
restriction fragments
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
restriction fragments

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
55
Match each term with its definition.
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
systems biology
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
systems biology

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
56
Match each term with its definition.
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
genetic engineering
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
genetic engineering

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
57
Match each term with its definition.
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
cDNA library
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
cDNA library

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
58
cDNA library is the partial collection of cloned cDNAs made from the mRNAs isolated from a cell.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
59
Match each term with its definition.
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
structural genomics
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
structural genomics

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
60
Match each term with its definition.
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
transgenic
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
transgenic

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
61
Match the names of the processes with the descriptions.
a.DNA fingerprinting
b.transgenics
c.DNA cloning
d.biotechnology
e.proteomics
This is the methodology that would be considered by a scientist who wants to generate mice that express a fluorescent protein in every cell of their body.
a.DNA fingerprinting
b.transgenics
c.DNA cloning
d.biotechnology
e.proteomics
This is the methodology that would be considered by a scientist who wants to generate mice that express a fluorescent protein in every cell of their body.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
62
Match each term with its definition.
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
genomic library
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
genomic library

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
63
Match the names of the processes to the descriptions. A process may be used once, more than once, or not at all.
a.DNA library
b.restriction enzyme
c.gene cloning
d.Southern blotting
e.PCR
You could use this technique to see if a DNA sequence is present in an individual's genome without having to use Sanger sequencing.
a.DNA library
b.restriction enzyme
c.gene cloning
d.Southern blotting
e.PCR
You could use this technique to see if a DNA sequence is present in an individual's genome without having to use Sanger sequencing.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
64
Match the names of the processes to the descriptions. A process may be used once, more than once, or not at all.
a.DNA library
b.restriction enzyme
c.gene cloning
d.Southern blotting
e.PCR
You would use this method to mass-produce a gene in a bacterial cell that does not normally have this gene.
a.DNA library
b.restriction enzyme
c.gene cloning
d.Southern blotting
e.PCR
You would use this method to mass-produce a gene in a bacterial cell that does not normally have this gene.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
65
Match the names of the processes with the descriptions.
a.DNA fingerprinting
b.transgenics
c.DNA cloning
d.biotechnology
e.proteomics
This is the technique that would be used by a forensic scientist trying to determine if a suspect is the actual rapist.
a.DNA fingerprinting
b.transgenics
c.DNA cloning
d.biotechnology
e.proteomics
This is the technique that would be used by a forensic scientist trying to determine if a suspect is the actual rapist.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
66
Match each term with its definition.
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
Southern blot
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
Southern blot

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
67
Match each term with its definition.
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
agarose gel electrophoresis
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
agarose gel electrophoresis

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
68
Match the names of the processes with the descriptions.
a.DNA fingerprinting
b.transgenics
c.DNA cloning
d.biotechnology
e.proteomics
This is any technique applied to living organisms that modifies or makes them for a specific purpose.
a.DNA fingerprinting
b.transgenics
c.DNA cloning
d.biotechnology
e.proteomics
This is any technique applied to living organisms that modifies or makes them for a specific purpose.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
69
Match the names of the processes to the descriptions. A process may be used once, more than once, or not at all.
a.DNA library
b.restriction enzyme
c.gene cloning
d.Southern blotting
e.PCR
You would use this method to determine the presence or absence of a particular 20 bp nucleotide pattern in a noncoding region of a genome.
a.DNA library
b.restriction enzyme
c.gene cloning
d.Southern blotting
e.PCR
You would use this method to determine the presence or absence of a particular 20 bp nucleotide pattern in a noncoding region of a genome.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
70
Match the names of the processes with the descriptions.
a.DNA fingerprinting
b.transgenics
c.DNA cloning
d.biotechnology
e.proteomics
This methodology lets you study the complete set of protein that can be expressed by an organism's genome.
a.DNA fingerprinting
b.transgenics
c.DNA cloning
d.biotechnology
e.proteomics
This methodology lets you study the complete set of protein that can be expressed by an organism's genome.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
71
Match the names of the processes to the descriptions. A process may be used once, more than once, or not at all.
a.DNA library
b.restriction enzyme
c.gene cloning
d.Southern blotting
e.PCR
You would use this to cut DNA at a selected site.
a.DNA library
b.restriction enzyme
c.gene cloning
d.Southern blotting
e.PCR
You would use this to cut DNA at a selected site.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
72
Match the names of the processes to the descriptions. A process may be used once, more than once, or not at all.
a.DNA library
b.restriction enzyme
c.gene cloning
d.Southern blotting
e.PCR
This technique is used to amplify specific sequences of DNA.
a.DNA library
b.restriction enzyme
c.gene cloning
d.Southern blotting
e.PCR
This technique is used to amplify specific sequences of DNA.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
73
Match each term with its definition.
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
DNA fingerprinting
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
DNA fingerprinting

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
74
Match each term with the most appropriate macromolecule being studied or manipulated.a.DNA
b.mRNA/cDNA
c.protein
blotting
b.mRNA/cDNA
c.protein
blotting

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
75
Match the names of the processes with the descriptions.
a.DNA fingerprinting
b.transgenics
c.DNA cloning
d.biotechnology
e.proteomics
You use this technique to produce recombinant DNA.
a.DNA fingerprinting
b.transgenics
c.DNA cloning
d.biotechnology
e.proteomics
You use this technique to produce recombinant DNA.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
76
Match each term with its definition.
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
recombinant DNA
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
recombinant DNA

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
77
Match each term with its definition.
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
PCR
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
PCR

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
78
Match each term with its definition.
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
proteomics
a.uses DNA technologies to alter genes in a cell or organism
b.branch of biology that tries to combine proteomics and genomics at an organismal level
c.determination and analysis of the nucleotide sequences in the chromosomes in an organism
d.products of a restriction endonuclease's action
e.collection of clones that together contain every DNA sequence in a genome, including noncoding sequences
f.use of a short single stranded DNA sequence, labelled with some sort of tag, to bind to complementary DNA and thus identify it
g.collection of clones that together contain a DNA version of every mRNA sequence expressed in a particular cell type
h.procedure described as "a photocopy machine for specific DNA sequences"
i.DNA from two different sources that have been joined together into a single molecule
j.term used to describe organisms that have been subject to genetic engineering
k.study of all of the proteins produced from an organism's genome
l.
study of the functions of genes and noncoding portions of the genome, including their interactions with each other
m.
bacterial enzymes that recognize and cut specific DNA sequences
n.
method of hybridizing labelled DNA to DNA fragments that were previously subjected to gel electrophoresis
o.
technique used to separate DNA molecules based on their relative sizes
p.
method of identifying individuals based on their individual patterns of short tandem repeats located on certain loci of their genome
proteomics

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
79
Match the names of the processes to the descriptions. A process may be used once, more than once, or not at all.
a.DNA library
b.restriction enzyme
c.gene cloning
d.Southern blotting
e.PCR
You would use this tool if you wanted to check if a particular gene was contained in an organism's genome.
a.DNA library
b.restriction enzyme
c.gene cloning
d.Southern blotting
e.PCR
You would use this tool if you wanted to check if a particular gene was contained in an organism's genome.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck
80
Match the names of the processes to the descriptions. A process may be used once, more than once, or not at all.
a.DNA library
b.restriction enzyme
c.gene cloning
d.Southern blotting
e.PCR
This technique will let you move a gene into a vector for expression in different cells.
a.DNA library
b.restriction enzyme
c.gene cloning
d.Southern blotting
e.PCR
This technique will let you move a gene into a vector for expression in different cells.

Unlock Deck
Unlock for access to all 91 flashcards in this deck.
Unlock Deck
k this deck


