Deck 5: The Generation of Lymphocyte Antigen Receptors
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/33
Play
Full screen (f)
Deck 5: The Generation of Lymphocyte Antigen Receptors
1
In rare instances, B cells can be found that have two immunoglobulin light chain alleles, both of which are rearranged in frame, and can encode functional light chain proteins. Yet, on the surface of the B cell, only one of the two light chain proteins is detected in the membrane-bound immunoglobulin receptor. The reason these rare cells have two functional light chain rearrangements but only express one of the two light chains as part of the B-cell receptor is:
A) One of the two light chains is formed from rearrangement of a V gene segment that is a pseudogene.
B) One of the two light chain proteins doesn't form a stable complex with the heavy chain expressed in this cell.
C) One of the two light chain alleles is not transcribed efficiently, and produces only low levels of protein.
D) One of the two light chain alleles uses a V gene segment that is not targeted very often by the RAG recombinase.
E) One of the two light chains is rapidly degraded after synthesis due to improper folding.
A) One of the two light chains is formed from rearrangement of a V gene segment that is a pseudogene.
B) One of the two light chain proteins doesn't form a stable complex with the heavy chain expressed in this cell.
C) One of the two light chain alleles is not transcribed efficiently, and produces only low levels of protein.
D) One of the two light chain alleles uses a V gene segment that is not targeted very often by the RAG recombinase.
E) One of the two light chains is rapidly degraded after synthesis due to improper folding.
One of the two light chain proteins doesn't form a stable complex with the heavy chain expressed in this cell.
2
B-cell receptors and T-cell receptors share a mechanism for generating diversity, and also share overall structural homology both in their V domains and their C domains. This is because the two proteins have nearly identical functions in the immune responses mediated by their respective cell types.
False
3
For immunoglobulin heavy and light chain genes, and for T-cell receptor chain genes, there are a large number of V gene segments, and relatively few J and/or D segments that rearrange to form the final coding sequence for each gene. The TCR locus is different in this regard, and this difference is thought to reflect the fact that nearly all : T-cell receptors recognize a peptide bound to an MHC molecule. This unique feature of the T-cell receptor locus is:
A) The presence of only five different V gene segments
B) The presence of two different C coding sequences
C) The presence of over sixty different J gene segments
D) The absence of D gene segments
E) The large sequence distance separating the V gene segments from the J gene segments
A) The presence of only five different V gene segments
B) The presence of two different C coding sequences
C) The presence of over sixty different J gene segments
D) The absence of D gene segments
E) The large sequence distance separating the V gene segments from the J gene segments
The presence of over sixty different J gene segments
4
An important mechanism for generating diversity in immunoglobulin light chain V-region sequences is based on the fact that the RAG recombinase generates hairpin structures, rather than blunt ends, at the cleavage sites between the recombination signal sequences and the coding sequences. Explain how this mechanism generates diversity at the junctions.

Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
5
For : T-cell receptors, sequence diversity is heavily concentrated at the junctions formed by the rearrangement of gene segments during the generation of the expressed V and V regions. The result of this organization is to position the most variable part of the T-cell receptor over a certain region of the ligand recognized by this receptor. Which region (outlined in red in Figure ) indicates this part of the ligand recognized by the T-cell receptor?
A)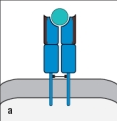
B)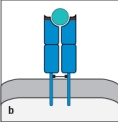
C)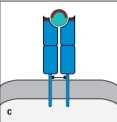
D)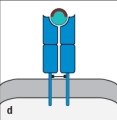
E)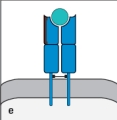
A)

B)

C)

D)

E)


Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
6
While B cells and T cells differ markedly in their functions during an immune response, the two lymphocyte subsets share the enzymatic machinery and overall scheme for generating antigen receptor diversity. This is because:
A) B cells and T cells recognize the same form of antigen expressed by an infecting pathogen.
B) Animals with B cells developed first, and later evolving species then developed T cells.
C) B cells and T cells both need enormous antigen receptor diversity to provide protection against the diversity of pathogens.
D) Antibody and T-cell receptor gene segments are both flanked by similar recombination signal sequences.
E) B cells and T cells both secrete their antigen receptor proteins after they are activated by antigen-binding.
A) B cells and T cells recognize the same form of antigen expressed by an infecting pathogen.
B) Animals with B cells developed first, and later evolving species then developed T cells.
C) B cells and T cells both need enormous antigen receptor diversity to provide protection against the diversity of pathogens.
D) Antibody and T-cell receptor gene segments are both flanked by similar recombination signal sequences.
E) B cells and T cells both secrete their antigen receptor proteins after they are activated by antigen-binding.

Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
7
Antibody diversity is generated by multiple mechanisms, each of which contributes to the generation of antibodies with up to 1011 different amino acid sequences in their antigen-binding sites. Several of these mechanisms involve changes in the DNA sequences encoding the antibody heavy and light chain proteins. One mechanism that does not rely on changes to the DNA within the immunoglobulin heavy and light chain gene loci is, instead, dependent on:
A) The contributions of amino acids from both the heavy chain and the light chain to form the antigen-binding site
B) The random usage of V, D, and J gene segments to form the heavy chain V region sequence
C) The random usage of light chains versus light chains to pair with the heavy chain
D) The activity of TdT to add random nucleotides at the junctions between the V, J, and D region sequences
E) The fact that heavy chain V regions contain an extra gene segment encoded by the D region compared to light chain V regions
A) The contributions of amino acids from both the heavy chain and the light chain to form the antigen-binding site
B) The random usage of V, D, and J gene segments to form the heavy chain V region sequence
C) The random usage of light chains versus light chains to pair with the heavy chain
D) The activity of TdT to add random nucleotides at the junctions between the V, J, and D region sequences
E) The fact that heavy chain V regions contain an extra gene segment encoded by the D region compared to light chain V regions

Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
8
Most of the enzymes involved in immunoglobulin gene rearrangement are ubiquitously expressed in all cells of the body. However, the specific recombination events between V, J, and D gene segments that generate antibody diversity occur only in developing B cells. How do RAG-1 and RAG-2 ensure that recombination takes place at antibody gene segments?

Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
9
The different classes of immunoglobulins differ in the sequences of their heavy chain constant regions. As a result, each class of antibody has distinct effector functions. Nonetheless, they are all found at about equal concentrations in the serum of healthy individuals.

Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
10
The exon encoding the V region of an immunoglobulin protein is generated by a process of somatic recombination. This recombination event brings V gene and J gene segments together:
A) In all cells of the body to encode a V region sequence
B) To generate maximum diversity in the CDR3 sequence of the V region
C) By alternative RNA splicing to encode a V region sequence
D) By a precise mechanism that never adds or loses nucleotides at the junction
E) To generate a single exon encoding the entire immunoglobulin protein
A) In all cells of the body to encode a V region sequence
B) To generate maximum diversity in the CDR3 sequence of the V region
C) By alternative RNA splicing to encode a V region sequence
D) By a precise mechanism that never adds or loses nucleotides at the junction
E) To generate a single exon encoding the entire immunoglobulin protein

Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
11
Most eukaryotic genes are encoded in a set of exons that are brought together to form a contiguous protein coding sequence by the process of mRNA splicing. In contrast, immunoglobulin genes use somatic recombination of gene segments and not mRNA splicing to generate the final mRNA that is translated into protein.

Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
12
When first discovered, investigators found it surprising that some single-gene defects causing immunodeficiency syndromes were associated with hypersensitivities to ionizing radiation, thereby leading to increased rates of cancer. The genes accounting for this dual impairment encode ubiquitously expressed DNA repair proteins.

Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
13
Antibodies that bind with high affinity to some viral surface proteins require heavy chain CDR3 loops of unusual length. Whereas the average human heavy chain CDR3 length is ~15 amino acids, antibodies with VH CDR3 loops of >30 amino acids are readily detected in the repertoire. These antibody heavy chains with CDR3 lengths of >30 amino acids would likely be missing in individuals lacking:
A) RAG-1 and RAG-2
B) DNA-dependent protein kinase (DNA-PK)
C) The nuclease, Artemis
D) The Ku70:Ku80 complex
E) Terminal deoxy nucleotidyl transferase (TdT)
A) RAG-1 and RAG-2
B) DNA-dependent protein kinase (DNA-PK)
C) The nuclease, Artemis
D) The Ku70:Ku80 complex
E) Terminal deoxy nucleotidyl transferase (TdT)

Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
14
Figure shows the germ-line configuration of three V gene segments (#1, 2, 3), and two J gene segments (#4, 5). Which of the choices below represents a DNA configuration that would result from V-to-J recombination?
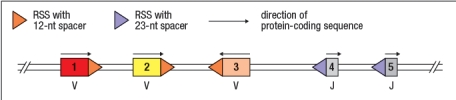
A)
B)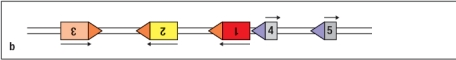
C)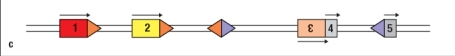
D)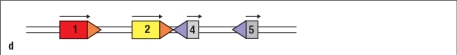
E)

A)

B)

C)

D)

E)


Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
15
Some pathogenic microorganisms encode proteins, such as the Staphylococcus Protein A, that bind to immunoglobulin constant region domains with high affinity. These microbial proteins provide a benefit to the microorganism by:
A) Preventing antibodies bound to the microbe from binding to Fc receptors on phagocytes
B) Blocking the binding of anti-microbial antibodies to the pathogen surface
C) Cleaving the antibody into fragments that separate the antigen-binding region from the effector function
D) Inducing aggregation of the anti-microbial antibodies by multivalent binding to the pathogen-derived protein
E) Preventing the antibody from neutralizing the pathogen
A) Preventing antibodies bound to the microbe from binding to Fc receptors on phagocytes
B) Blocking the binding of anti-microbial antibodies to the pathogen surface
C) Cleaving the antibody into fragments that separate the antigen-binding region from the effector function
D) Inducing aggregation of the anti-microbial antibodies by multivalent binding to the pathogen-derived protein
E) Preventing the antibody from neutralizing the pathogen

Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
16
Recombination signal sequences are conserved heptamer and nonamer sequences that flank the V, J, and D gene segments which undergo recombination to generate the final V region coding exon. Some of these have 12-nucleotide spacers between the heptamer and nonamer, and others have 23-nucleotide spacers. The reason recombination signal sequences come in these two forms is:
A) To ensure the correct assembly of gene segments so that a VH recombines to a DH and not to another VH, for instance
B) To ensure that the heptamer and nonamer are found on the same face of the DNA double helix
C) To ensure that , , and heavy chains recombine within a locus and not between loci
D) To ensure that , , and heavy chain gene segments do not undergo recombination with non-immunoglobulin genes
E) To ensure that the RAG recombinase cuts the DNA between the last nucleotide of the heptamer and the coding sequence
A) To ensure the correct assembly of gene segments so that a VH recombines to a DH and not to another VH, for instance
B) To ensure that the heptamer and nonamer are found on the same face of the DNA double helix
C) To ensure that , , and heavy chains recombine within a locus and not between loci
D) To ensure that , , and heavy chain gene segments do not undergo recombination with non-immunoglobulin genes
E) To ensure that the RAG recombinase cuts the DNA between the last nucleotide of the heptamer and the coding sequence

Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
17
Different individuals can have different numbers of functional V gene segments as well as different numbers of constant region genes. This type of genetic polymorphism between individuals indicates that:
A) The antibody heavy and light chain loci undergo more frequent mutation than other genes in the genome.
B) The recombination machinery is active in germ cells.
C) Individuals only need or light chains, but not both.
D) The precise number of antibody gene segments in an individual is not important.
E) Antibody gene segments underwent more frequent duplication during evolution than other genes in the genome.
A) The antibody heavy and light chain loci undergo more frequent mutation than other genes in the genome.
B) The recombination machinery is active in germ cells.
C) Individuals only need or light chains, but not both.
D) The precise number of antibody gene segments in an individual is not important.
E) Antibody gene segments underwent more frequent duplication during evolution than other genes in the genome.

Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
18
Some T cells express : T-cell receptors rather than : T-cell receptors. The organization of the locus and the locus helps to ensure that each T cell cannot express both types of T-cell receptors. The mechanism involved is that:
A) The rearrangement of a T-cell receptor gene deletes the locus on that allele.
B) The rearrangement of a T-cell receptor gene deletes the locus on that allele.
C) The RAG recombinase enzymes are down-regulated immediately after the first T-cell receptor genes rearrange.
D) The : T-cell receptor signals the T cell to delete the locus.
E) The : T-cell receptor signals the T cell to delete the locus.
A) The rearrangement of a T-cell receptor gene deletes the locus on that allele.
B) The rearrangement of a T-cell receptor gene deletes the locus on that allele.
C) The RAG recombinase enzymes are down-regulated immediately after the first T-cell receptor genes rearrange.
D) The : T-cell receptor signals the T cell to delete the locus.
E) The : T-cell receptor signals the T cell to delete the locus.

Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
19
IgM is the first antibody isotype secreted following activation of a naive B cell. IgM is found at high concentrations in the serum, and is found as a very high molecular weight complex. This high molecular weight complex is composed of:
A) A single IgM monomer plus monomers of IgA and IgG
B) A single IgM monomer bound to several non-immunoglobulin serum proteins
C) A single IgM monomer bound to serum complement components
D) A pentamer of IgM monomers
E) Two dimers of IgM plus IgD forming a tetrameric complex
A) A single IgM monomer plus monomers of IgA and IgG
B) A single IgM monomer bound to several non-immunoglobulin serum proteins
C) A single IgM monomer bound to serum complement components
D) A pentamer of IgM monomers
E) Two dimers of IgM plus IgD forming a tetrameric complex

Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
20
The addition and subtraction of nucleotides at the junctions between V, D, and J gene segments creates antibody proteins with wide variations in the numbers of amino acids in their CDR3 regions. This variability in CDR3 length is important as:
A) Overall variability in CDR3 sequence is needed to create a sufficiently diverse antibody repertoire.
B) The CDR3 region is more important in binding antigen than the CDR1 and CDR2 regions are.
C) Some light chains bind better to heavy chains with longer CDR3 region sequences.
D) Longer CDR3 sequences generally create antibodies with higher affinity for the antigen.
E) Some antibodies bind relatively flat surfaces and others bind deep clefts in the antigen.
A) Overall variability in CDR3 sequence is needed to create a sufficiently diverse antibody repertoire.
B) The CDR3 region is more important in binding antigen than the CDR1 and CDR2 regions are.
C) Some light chains bind better to heavy chains with longer CDR3 region sequences.
D) Longer CDR3 sequences generally create antibodies with higher affinity for the antigen.
E) Some antibodies bind relatively flat surfaces and others bind deep clefts in the antigen.

Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
21
The V-D-J recombination process used to generate antigen receptor diversity in vertebrates is believed to have evolved from a transposon. How does this hypothesis explain why the recombination signal sequences of the antigen receptor gene segments are joined precisely during the recombination process, whereas the cut ends that are joined to form the antigen receptor coding sequence are joined by an error-prone process?

Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
22
Several invertebrate species, such as some insect and snail species, have mechanisms for increasing the diversity of immune recognition molecules that are expressed in an individual beyond the simple 'one gene encodes one protein' rule that applies to most genes in the genome. One of these mechanisms:
A) Uses a pair of enzymes that are related to the RAG recombinase enzymes found in vertebrates
B) Creates a library of complement proteins that function like vertebrate complement to promote phagocytosis of microbes by macrophages and neutrophils
C) Creates a clonally distributed set of immune recognition molecules by a process involving different DNA rearrangement events in each cell
D) Uses a mechanism in which a small number of cells expressing a few different proteins undergo massive proliferation to provide protective immunity
E) Resembles the V-D-J recombination process in vertebrates but occurs by differential RNA splicing rather than by DNA rearrangement
A) Uses a pair of enzymes that are related to the RAG recombinase enzymes found in vertebrates
B) Creates a library of complement proteins that function like vertebrate complement to promote phagocytosis of microbes by macrophages and neutrophils
C) Creates a clonally distributed set of immune recognition molecules by a process involving different DNA rearrangement events in each cell
D) Uses a mechanism in which a small number of cells expressing a few different proteins undergo massive proliferation to provide protective immunity
E) Resembles the V-D-J recombination process in vertebrates but occurs by differential RNA splicing rather than by DNA rearrangement

Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
23
Classical MHC molecules function as peptide-binding receptors that present these peptides to : T cells for recognition by : T-cell receptors. MHC molecules can be detected as far back in evolution as sharks, the same time at which T-cell receptors can be identified. Examination of shark MHC proteins indicates that these molecules likely function identically to human MHC molecules. This conclusion is based on:
A) The conservation of amino acid residues important in peptide binding in both shark and human MHC proteins
B) The observation that MHC genes are highly polymorphic in sharks as well as humans
C) The fact that sharks have both MHC class I and MHC class II molecules
D) The finding that sharks have both classical and nonclassical MHC molecules, like humans
E) The ability to trace the evolution of MHC protein sequences from the earliest vertebrates to humans
A) The conservation of amino acid residues important in peptide binding in both shark and human MHC proteins
B) The observation that MHC genes are highly polymorphic in sharks as well as humans
C) The fact that sharks have both MHC class I and MHC class II molecules
D) The finding that sharks have both classical and nonclassical MHC molecules, like humans
E) The ability to trace the evolution of MHC protein sequences from the earliest vertebrates to humans

Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
24
In some vertebrates, such as rays and some shark species, immunoglobulin light chain genes consist of multiple units of already rearranged VJ-C genes, of which one is chosen for expression in a developing B cell. This strategy may have evolved in these organisms:
A) Prior to the evolution of rearranging gene segments requiring somatic recombination in developing B cells
B) Prior to the evolution of the RAG1 and RAG2 enzymes needed for recombination
C) As a mechanism to increase light chain diversity beyond that achieved by the use of rearranging gene segments in humans
D) As a mechanism to increase light chain diversity beyond that achieved the gene conversion process used in chickens
E) As a means to generate a rapid response to common pathogens encountered by these animals
A) Prior to the evolution of rearranging gene segments requiring somatic recombination in developing B cells
B) Prior to the evolution of the RAG1 and RAG2 enzymes needed for recombination
C) As a mechanism to increase light chain diversity beyond that achieved by the use of rearranging gene segments in humans
D) As a mechanism to increase light chain diversity beyond that achieved the gene conversion process used in chickens
E) As a means to generate a rapid response to common pathogens encountered by these animals

Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
25
A mutagenesis screen performed on mice identified a gene with an important function in B cells. Analysis of B cells from the spleens of these mutant mice showed the results shown in Figure. However, when the C coding sequences were determined, no mutations in these DNA sequences were found. To study this further, genetic crosses were set-up with another mouse strain carrying an allelic variant of the immunoglobulin heavy chain locus. The original mutant mouse strain (strain A) carries IgMa and IgDa alleles of these heavy chain genes. The second mouse strain (strain B) carries the IgMb and IgDb alleles of the heavy chain genes. These two parental strains were crossed together, generating progeny that were all heterozygous for IgM and IgD alleles B were obtained. 
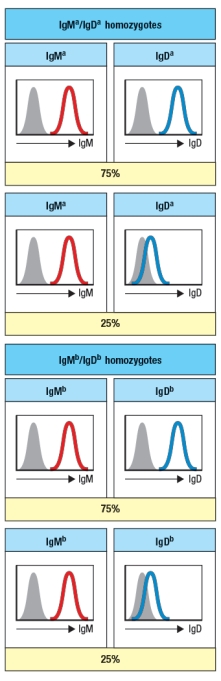
Based on these data, the mutation in the original mutant mouse strain most likely inactivates:
A) The gene for a transcription factor required for IgD gene transcription
B) A regulatory region in the immunoglobulin heavy chain gene locus required for IgD gene transcription
C) The gene for a factor required for alternative mRNA splicing of the immunoglobulin heavy chain primary mRNA transcript
D) The gene encoding the RAG-1 or RAG-2 recombinase
E) The gene for a factor that is required for IgD surface expression


Based on these data, the mutation in the original mutant mouse strain most likely inactivates:
A) The gene for a transcription factor required for IgD gene transcription
B) A regulatory region in the immunoglobulin heavy chain gene locus required for IgD gene transcription
C) The gene for a factor required for alternative mRNA splicing of the immunoglobulin heavy chain primary mRNA transcript
D) The gene encoding the RAG-1 or RAG-2 recombinase
E) The gene for a factor that is required for IgD surface expression

Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
26
When a B cell differentiates into a plasma cell, it goes from expressing membrane-bound IgM to making mostly the secreted form of IgM. A deletion of the first polyadenylation site in the μ heavy chain gene would prevent activated B cells from making the secreted form of IgM.

Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
27
Immunodeficiency diseases arise when individuals lack one or more components of their immune system, and are identified by an individual's history of persistent or recurrent infections. Some genetic defects (mutations or small deletions) can cause profound defects in an immune cell population; alternatively, in some cases, such small defects occur that there is no visible effect on immune responses. The diagram in Figure shows simplified versions of the immunoglobulin heavy chain locus, the T-cell receptor chain locus, and the locus encoding the RAG-1 and RAG-2 recombinases. For the sake of this question, imagine that these diagrams represent all of the gene segments present in the immunoglobulin heavy chain and T-cell receptor chain locus. 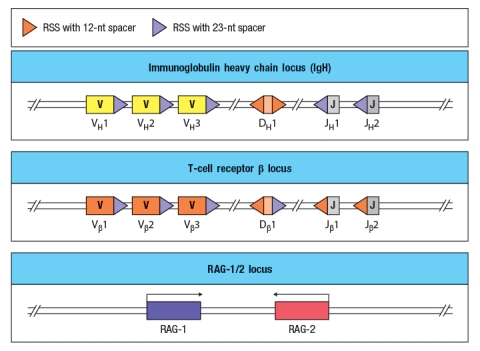
You now analyze five individuals, each of which has a single inactivating mutation in a region of one of these three loci. These mutations are each indicated by a red 'X' in Figure , and are numbered 1-5. For each of these inactivating mutations, indicate the alterations and/or defects that would be seen in the repertoire of antigen receptors found in mature B and T cells in that individual. Also, for each mutation, indicate whether the individual would likely show any immunodeficiency, such as a history of recurrent infections.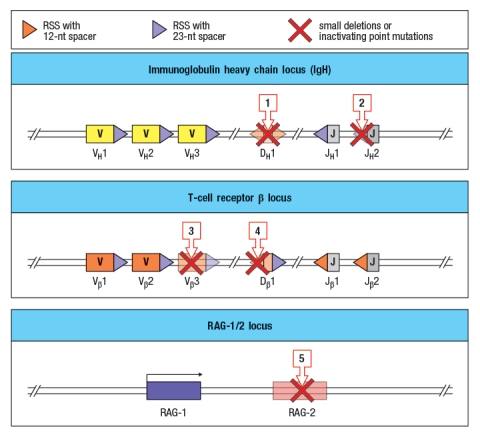

You now analyze five individuals, each of which has a single inactivating mutation in a region of one of these three loci. These mutations are each indicated by a red 'X' in Figure , and are numbered 1-5. For each of these inactivating mutations, indicate the alterations and/or defects that would be seen in the repertoire of antigen receptors found in mature B and T cells in that individual. Also, for each mutation, indicate whether the individual would likely show any immunodeficiency, such as a history of recurrent infections.


Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
28
The variable lymphocyte receptors (VLRs) of lampreys represent a highly diverse set of proteins with structural similarity to the mammalian Toll-like receptors. Yet the VLRs and the lymphocytes that produce them are thought to be components of the lamprey's adaptive immune system. This is because:
A) The VLRs are highly diverse and have been shown to bind to pathogens.
B) Both membrane-bound and secreted forms of VLRs are found in the lampreys.
C) VLR coding genes are formed by DNA rearrangement events.
D) VLRs are clonally distributed such that each lymphocyte expresses only one form of VLR.
E) VLR gene rearrangements occur by a process similar to gene conversion.
A) The VLRs are highly diverse and have been shown to bind to pathogens.
B) Both membrane-bound and secreted forms of VLRs are found in the lampreys.
C) VLR coding genes are formed by DNA rearrangement events.
D) VLRs are clonally distributed such that each lymphocyte expresses only one form of VLR.
E) VLR gene rearrangements occur by a process similar to gene conversion.

Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
29
In spite of their low affinity for binding to their antigen, IgM antibodies can be quite effective at promoting the elimination of extracellular bacterial infections. This is due to:
A) The ability of IgM antibodies to traffic across epithelial surfaces
B) Multivalent binding of IgM pentamers to repetitive epitopes on bacterial surfaces
C) The high concentration of IgM antibodies in the serum and in all tissues
D) The early production of IgM antibodies during an immune response
E) The high rate of somatic hypermutation in the genes encoding IgM antibodies
A) The ability of IgM antibodies to traffic across epithelial surfaces
B) Multivalent binding of IgM pentamers to repetitive epitopes on bacterial surfaces
C) The high concentration of IgM antibodies in the serum and in all tissues
D) The early production of IgM antibodies during an immune response
E) The high rate of somatic hypermutation in the genes encoding IgM antibodies

Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
30
T-cell receptor spectratype analysis is used to examine the diversity of T-cell receptor chain sequences in an individual's T cells. For this technique, T cells are isolated from a sample of thymocytes (developing T cells) or mature peripheral T cells from an individual. The mRNA is isolated from these cells and cDNA is generated by reverse transcription. This pool of cDNA is mixed with PCR primers that are used to amplify part of the rearranged T-cell receptor chain sequence containing the complete CDR3. The position of these primers relative to the rearranged T-cell receptor chain gene in the DNA locus is shown in Figure. Following the PCR amplification, the heterogeneous mixture of DNA molecules is then size-separated by electrophoresis on an apparatus that can separate molecules that differ by a single nucleotide. At the end, the quantity of material deposited in each band of a given nucleotide sequence length is quantified by densitometry, and the spectratype trace is produced. The x-axis of the spectratype depicts the number of nucleotides in each PCR product from the beginning of the forward primer to the end of the reverse primer.
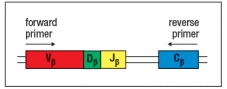
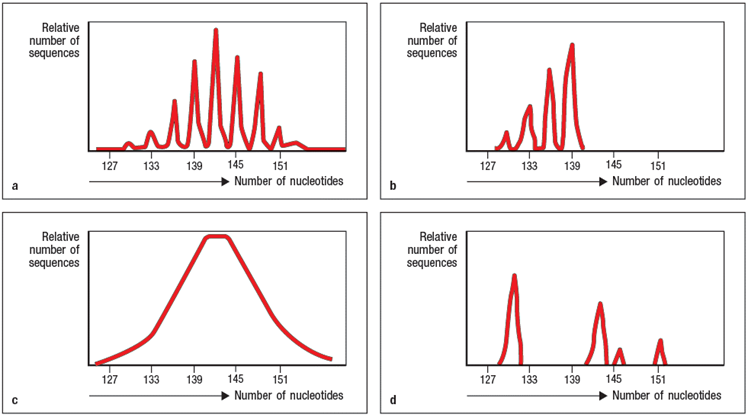
a) Panel A of Figure shows the spectratype trace of mature peripheral T cells from a healthy individual. What is explanation for the separation of the heterogeneous population of T-cell receptor chain sequences into multiple sharp peaks of size lengths?
b) Panel B shows T cells from an individual that is missing an important enzyme that contributes to T-cell receptor chain diversity during the recombination process. Which enzyme is most likely absent in this individual?
c) Panel C shows the spectratype analysis of T-cell receptor chain sequences in developing T cells that have just completed the V-D-J recombination process. Explain why this spectratype looks different from the one shown in panel A.
d) Panel D shows a more restricted example of spectratype analysis, where the forward primer used only binds to one specific V sequence. In this example, the primer is specific for V 17. When such a V -specific primer is used, the spectratype analysis only shows the junctional sequence lengths for T cells whose chain uses V 17. In a healthy individual, the V 17 spectratype would like just like the one shown in panel A; in other words, it would show a random distribution of V 17+ T-cell receptor chains with a normal distribution of junctional lengths.
However, in this case, the individual being studied has been infected with influenza virus, and is in the midst of a robust T cell response against the virus. What is the likely explanation for the non-random pattern of peaks on the V 17 spectratype from this individual at this timepoint?


a) Panel A of Figure shows the spectratype trace of mature peripheral T cells from a healthy individual. What is explanation for the separation of the heterogeneous population of T-cell receptor chain sequences into multiple sharp peaks of size lengths?
b) Panel B shows T cells from an individual that is missing an important enzyme that contributes to T-cell receptor chain diversity during the recombination process. Which enzyme is most likely absent in this individual?
c) Panel C shows the spectratype analysis of T-cell receptor chain sequences in developing T cells that have just completed the V-D-J recombination process. Explain why this spectratype looks different from the one shown in panel A.
d) Panel D shows a more restricted example of spectratype analysis, where the forward primer used only binds to one specific V sequence. In this example, the primer is specific for V 17. When such a V -specific primer is used, the spectratype analysis only shows the junctional sequence lengths for T cells whose chain uses V 17. In a healthy individual, the V 17 spectratype would like just like the one shown in panel A; in other words, it would show a random distribution of V 17+ T-cell receptor chains with a normal distribution of junctional lengths.
However, in this case, the individual being studied has been infected with influenza virus, and is in the midst of a robust T cell response against the virus. What is the likely explanation for the non-random pattern of peaks on the V 17 spectratype from this individual at this timepoint?

Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
31
In some cases, antibody binding to a pathogen will prevent the pathogen from adhering to and/or from infecting host cells. This antibody function does not require the Fc region of the antibody. Nonetheless, the Fc region is still required to facilitate the removal of the antibody:pathogen complex from the body. Explain why.

Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
32
The generation of a complete coding sequence for an antibody heavy chain involves a lymphocyte-restricted process of DNA rearrangement that links V, D, and J gene segments together to form the exon that encodes the heavy chain V region. A similar type of DNA rearrangement is also utilized for the simultaneous expression of IgM and IgD antibodies by the same B cell.

Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck
33
The evolutionary conservation of two classes of T lymphocytes, those expressing : T-cell receptors and those expressing : T-cell receptors, argues for important but separate functions of these two classes of T cells. Evolution has also conserved the process of somatic recombination of gene segments that is able to generate an enormous diversity of : and : T-cell receptor sequences. Do these observations indicate that both classes of T cells are components of the adaptive immune system? Why or why not?

Unlock Deck
Unlock for access to all 33 flashcards in this deck.
Unlock Deck
k this deck


