Deck 6: Antigen Presentation to T Lymphocytes
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/30
Play
Full screen (f)
Deck 6: Antigen Presentation to T Lymphocytes
1
MHC class I surface expression is dependent on an abundant source of pathogen-derived peptides. Thus, in uninfected cells, nearly all of the MHC class I proteins are degraded and never reach the cell surface.
False
2
Multiple mechanisms contribute to create a wide diversity in MHC protein expression between different individuals in the population. In addition to the genetic polymorphism of MHC genes, what additional mechanism(s) contribute to this diversity?
Polygeny and co-dominant expression of MHC proteins.
The high polymorphism of the classical MHC genes ensures diversity in MHC gene expression in the population as a whole. However, no matter how polymorphic a gene is, no individual can express more than two alleles at a single gene locus. Polygeny, the presence of several different related genes with similar functions, along with the co-dominant expression of both alleles of each MHC gene, ensures that each individual produces a number of different MHC molecules. The combination of polymorphism, polygeny, and co-dominant expression produces the diversity of MHC molecules seen both within an individual and in the population at large.
The high polymorphism of the classical MHC genes ensures diversity in MHC gene expression in the population as a whole. However, no matter how polymorphic a gene is, no individual can express more than two alleles at a single gene locus. Polygeny, the presence of several different related genes with similar functions, along with the co-dominant expression of both alleles of each MHC gene, ensures that each individual produces a number of different MHC molecules. The combination of polymorphism, polygeny, and co-dominant expression produces the diversity of MHC molecules seen both within an individual and in the population at large.
3
Three major cell types, dendritic cells, macrophages, and B cells, present peptides bound to MHC class II molecules for recognition by CD4 T cells. In general, these peptides are derived from proteins or pathogens taken up by the cell by endocytosis, phagocytosis, or macropinocytosis. Based on these pathways of antigen uptake, some of the enzymes that degrade proteins to generate peptides for MHC class II presentation are:
A) Ubiquitin ligases that tag proteins for degradation by the proteasome
B) ATP transporter proteins that deliver endocytic proteins into the cytosol for degradation
C) Cysteine proteases like cathepsins that function at acidic pH
D) The lysosomal thiol reductase found in the endosomes
E) The lysosome-associated membrane trafficking protein, LAMP-2
A) Ubiquitin ligases that tag proteins for degradation by the proteasome
B) ATP transporter proteins that deliver endocytic proteins into the cytosol for degradation
C) Cysteine proteases like cathepsins that function at acidic pH
D) The lysosomal thiol reductase found in the endosomes
E) The lysosome-associated membrane trafficking protein, LAMP-2
Cysteine proteases like cathepsins that function at acidic pH
4
The experiment shown in Figure uses two strains of mice that differ in their MHC genes. Strain A is H-2a and Strain B is H-2b. Mice of each strain are infected with the virus LCMV, and T cells are isolated at day 8 post-infection. These T cells are mixed with target cells that express either H-2a or H-2b; in each case, the target cells are either uninfected or infected with LCMV. After a four-hour incubation of T cells with target cells, the percentage of target cells lysed by the T cells is shown in the graph. 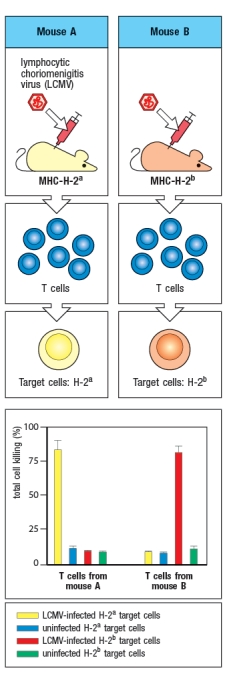 The explanation for the results of this experiment is:
The explanation for the results of this experiment is:
A) Mice of strain B do not make a T cell response to LCMV.
B) Mice of strain A make a more robust T cell response to LCMV than mice of strain B.
C) Target cells that express H-2b cannot be infected with LCMV.
D) T cells from mice of strain A only recognize viral peptides on target cells expressing H-2a.
E) LCMV peptides do not bind to MHC class I molecules from H-2b mice.
 The explanation for the results of this experiment is:
The explanation for the results of this experiment is:A) Mice of strain B do not make a T cell response to LCMV.
B) Mice of strain A make a more robust T cell response to LCMV than mice of strain B.
C) Target cells that express H-2b cannot be infected with LCMV.
D) T cells from mice of strain A only recognize viral peptides on target cells expressing H-2a.
E) LCMV peptides do not bind to MHC class I molecules from H-2b mice.

Unlock Deck
Unlock for access to all 30 flashcards in this deck.
Unlock Deck
k this deck
5
The MARCH-1 E3-ubiquitin ligase is expressed in B cells, dendritic cells, and macrophages. The pathway regulated by MARCH-1 is targeted by some pathogens in an immune evasion strategy. In this strategy, the pathogens encode:
A) A protein that induces degradation of MARCH-1
B) A protein that mimics MARCH-1 and functions similarly
C) A protein that binds to MARCH-1 and inhibits its function
D) A protein that is induced by IL-10 in macrophages and dendritic cells
E) A protein that induces degradation of CD86
A) A protein that induces degradation of MARCH-1
B) A protein that mimics MARCH-1 and functions similarly
C) A protein that binds to MARCH-1 and inhibits its function
D) A protein that is induced by IL-10 in macrophages and dendritic cells
E) A protein that induces degradation of CD86

Unlock Deck
Unlock for access to all 30 flashcards in this deck.
Unlock Deck
k this deck
6
In a mixed lymphocyte reaction, T cells from individual A make a robust response to antigen-presenting-cells from individual B, as long as the two individuals express different alleles of MHC molecules. Estimates indicate that up to 10% of the T cells from individual A may contribute to this response. If one performed this assay using responder T cells from a child and antigen-presenting cells from one parent, the result would be:
A) A massive proliferative response made by the antigen-presenting cells of the parent
B) A very weak response by the child's T cells, involving only 0.1% of their T cells
C) The complete absence of any proliferative response by the child's T cells
D) A robust cytolytic response that kills all of the parent's antigen-presenting cells
E) A robust response by the child's T cells
A) A massive proliferative response made by the antigen-presenting cells of the parent
B) A very weak response by the child's T cells, involving only 0.1% of their T cells
C) The complete absence of any proliferative response by the child's T cells
D) A robust cytolytic response that kills all of the parent's antigen-presenting cells
E) A robust response by the child's T cells

Unlock Deck
Unlock for access to all 30 flashcards in this deck.
Unlock Deck
k this deck
7
The mechanism of cross-presentation by dendritic cells is an essential pathway for generating CD8 T cell responses to some intracellular pathogens. If this pathway did not exist, we would be highly susceptible to:
A) Intracellular pathogens that can survive inside macrophage endocytic vesicles
B) Intracellular pathogens that are able to evade antibody responses
C) Intracellular pathogens that do not infect and replicate in dendritic cells
D) Intracellular pathogens that can spread from cell to cell by inducing cell fusion
E) Intracellular pathogens that infect and replicate in red blood cells
A) Intracellular pathogens that can survive inside macrophage endocytic vesicles
B) Intracellular pathogens that are able to evade antibody responses
C) Intracellular pathogens that do not infect and replicate in dendritic cells
D) Intracellular pathogens that can spread from cell to cell by inducing cell fusion
E) Intracellular pathogens that infect and replicate in red blood cells

Unlock Deck
Unlock for access to all 30 flashcards in this deck.
Unlock Deck
k this deck
8
Virus infections induce production of interferons that act on infected cells to enhance their recognition by CD8 cytotoxic T cells. To counter these mechanisms, viruses often encode proteins that interfere with antigen processing and presentation. In an experiment, cells infected with Virus X are treated with interferon and compared with uninfected cells treated with interferon. Proteasomes are isolated from the two cell populations and their enzymatic activities are compared. The data in Figure show the amino acid preferences for cleavage of peptides by the two samples of proteasomes. 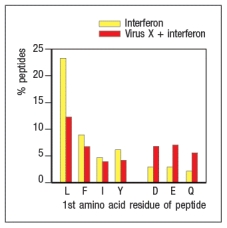
Based on these data, Virus X most likely encodes a protein that interferes with:
A) The expression of MHC class I on the surface of the infected cell
B) The rate at which peptides are produced from intact proteins in the infected cell
C) The transport of peptides from the cytosol to the endoplasmic reticulum in the infected cell
D) The replacement of constitutive proteasome subunits with immunoproteasome subunits in the infected cell
E) The development of CD8 T cells in the thymus by inhibiting the thymoproteasome

Based on these data, Virus X most likely encodes a protein that interferes with:
A) The expression of MHC class I on the surface of the infected cell
B) The rate at which peptides are produced from intact proteins in the infected cell
C) The transport of peptides from the cytosol to the endoplasmic reticulum in the infected cell
D) The replacement of constitutive proteasome subunits with immunoproteasome subunits in the infected cell
E) The development of CD8 T cells in the thymus by inhibiting the thymoproteasome

Unlock Deck
Unlock for access to all 30 flashcards in this deck.
Unlock Deck
k this deck
9
The virus shown in the diagram below is only able to infect and replicate in epithelial cells. In order for the cross-presenting dendritic cell to display viral peptides, rather than self peptides on its surface MHC class I proteins, which of the following procedures could be utilized, starting with the components shown in Figure? 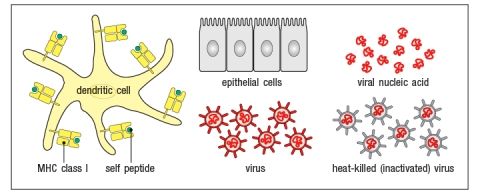
A) Mix epithelial cells with heat-killed virus, wait 24 hrs, wash away any virus particles outside the epithelial cells, then add epithelial cells to dendritic cells.
B) Mix epithelial cells with viral peptides, wait 24 hrs, wash away any viral peptides not bound to the epithelial cells, then add epithelial cells to dendritic cells.
C) Mix epithelial cells with live virus particles, wait 24 hrs, wash away any virus particles outside the epithelial cells, then add epithelial cells to dendritic cells.
D) Mix dendritic cells with viral nucleic acids and epithelial cells for 24 hrs.
E) MIx epithelial cells will viral nucleic acids, wait 24 hrs, wash away any viral nucleic acid remaining outside the epithelial cells, then add epithelial cells to dendritic cells.

A) Mix epithelial cells with heat-killed virus, wait 24 hrs, wash away any virus particles outside the epithelial cells, then add epithelial cells to dendritic cells.
B) Mix epithelial cells with viral peptides, wait 24 hrs, wash away any viral peptides not bound to the epithelial cells, then add epithelial cells to dendritic cells.
C) Mix epithelial cells with live virus particles, wait 24 hrs, wash away any virus particles outside the epithelial cells, then add epithelial cells to dendritic cells.
D) Mix dendritic cells with viral nucleic acids and epithelial cells for 24 hrs.
E) MIx epithelial cells will viral nucleic acids, wait 24 hrs, wash away any viral nucleic acid remaining outside the epithelial cells, then add epithelial cells to dendritic cells.

Unlock Deck
Unlock for access to all 30 flashcards in this deck.
Unlock Deck
k this deck
10
Some viruses have mechanisms to down-regulate MHC class I protein expression on the surface of cells in which the virus is replicating. This immune evasion strategy might prevent effector CD8 cytotoxic T cells from recognizing and killing the virus-infected cells. Would this immune evasion strategy also prevent the initial activation of virus-specific CD8 T cells?
A) Yes, because no viral peptide:MHC class I complexes would form to activate CD8 T cells.
B) No, because dendritic cells would take up infected cells and cross-present viral peptides to activate CD8 T cells.
C) No, because some presentation of MHC class I complexes with viral peptides would occur before the virus could down-regulate all the surface MHC class I protein.
D) Yes, because this immune evasion strategy would also function in dendritic cells, even if the virus doesn't replicate in dendritic cells.
E) No, because the type I interferon response induced by the virus infection will up-regulate MHC class I expression and override the immune evasion mechanism.
A) Yes, because no viral peptide:MHC class I complexes would form to activate CD8 T cells.
B) No, because dendritic cells would take up infected cells and cross-present viral peptides to activate CD8 T cells.
C) No, because some presentation of MHC class I complexes with viral peptides would occur before the virus could down-regulate all the surface MHC class I protein.
D) Yes, because this immune evasion strategy would also function in dendritic cells, even if the virus doesn't replicate in dendritic cells.
E) No, because the type I interferon response induced by the virus infection will up-regulate MHC class I expression and override the immune evasion mechanism.

Unlock Deck
Unlock for access to all 30 flashcards in this deck.
Unlock Deck
k this deck
11
The adaptive immune system uses multiple strategies to generate diversity in our ability to mount responses to a wide array of infectious microorganisms. These strategies include the generation of diverse repertoires of B-cell and T-cell antigen receptors, as well as polymorphism of MHC genes. The polymorphism of MHC genes differs from the diversity of lymphocyte antigen receptors in that:
A) It involves DNA rearrangements at multiple gene segments in the MHC locus.
B) It requires different enzymes than the RAG1/RAG2 recombinase required for antigen receptor rearrangements.
C) It results in a diverse repertoire of clonally distributed receptors on dendritic cells, rather than on lymphocytes.
D) It creates diversity between individuals in the population rather than within a single individual.
E) It does not contribute to the transplant rejection responses that occur after organ transplantation between unrelated individuals.
A) It involves DNA rearrangements at multiple gene segments in the MHC locus.
B) It requires different enzymes than the RAG1/RAG2 recombinase required for antigen receptor rearrangements.
C) It results in a diverse repertoire of clonally distributed receptors on dendritic cells, rather than on lymphocytes.
D) It creates diversity between individuals in the population rather than within a single individual.
E) It does not contribute to the transplant rejection responses that occur after organ transplantation between unrelated individuals.

Unlock Deck
Unlock for access to all 30 flashcards in this deck.
Unlock Deck
k this deck
12
The invariant chain protein, Ii, has only one function in MHC class II antigen presentation. This function entails Ii protein occupying the peptide-binding site of each newly synthesized class II protein, thereby preventing nascent MHC class II proteins from binding peptides or misfolded proteins in the endoplasmic reticulum.

Unlock Deck
Unlock for access to all 30 flashcards in this deck.
Unlock Deck
k this deck
13
MHC polymorphism at individual MHC genes appears to have been strongly selected by evolutionary pressures. In other words, there appears to be selection for maintaining hundreds to thousands of different alleles of each MHC gene in the population. This notion is based on the observation that nucleotide differences between alleles that lead to amino acid substitutions are more frequent than those that are silent substitutions (i.e., not changing the amino acid sequence of the protein). In addition, the positions within the MHC protein where most of the allelic sequence variation occurs are not randomly distributed, but are concentrated in certain regions of the MHC protein. This latter point indicates:
A) That some nucleotide sequences within the MHC genes are hot-spots for mutation
B) That MHC genes are more susceptible to point mutations than to larger nucleotide deletions
C) That MHC allelic polymorphism has been driven by selection for diversity in peptide binding specificity
D) That MHC genes are more susceptible to all types of mutations than are other genes in the genome
E) That MHC polymorphism has evolved to prevent pathogens that infect non-human primates from infecting humans
A) That some nucleotide sequences within the MHC genes are hot-spots for mutation
B) That MHC genes are more susceptible to point mutations than to larger nucleotide deletions
C) That MHC allelic polymorphism has been driven by selection for diversity in peptide binding specificity
D) That MHC genes are more susceptible to all types of mutations than are other genes in the genome
E) That MHC polymorphism has evolved to prevent pathogens that infect non-human primates from infecting humans

Unlock Deck
Unlock for access to all 30 flashcards in this deck.
Unlock Deck
k this deck
14
The diagram in Figure shows a pathogen (in red) that is present in different cellular compartments of each of the cell types shown. In each case, a specific T cell subset will recognize peptides of that pathogen presented on MHC molecules on the surface of the cell, and will execute its effector function. From the list below, match the appropriate T cell effector response to the cell type and location of the pathogen. 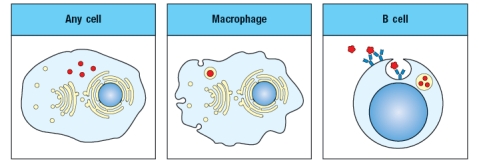
i. CD4 T cell killing of target cell
ii. CD8 T cell killing of target cell
iii. CD4 T cell activation of target cell's antibody production
iv. CD8 T cell activation of target cell's antibody production
v. CD4 T cell activation of target cell's ability to kill intracellular pathogen
vi. CD8 T cell activation of target cell's ability to kill intracellular pathogen

i. CD4 T cell killing of target cell
ii. CD8 T cell killing of target cell
iii. CD4 T cell activation of target cell's antibody production
iv. CD8 T cell activation of target cell's antibody production
v. CD4 T cell activation of target cell's ability to kill intracellular pathogen
vi. CD8 T cell activation of target cell's ability to kill intracellular pathogen

Unlock Deck
Unlock for access to all 30 flashcards in this deck.
Unlock Deck
k this deck
15
The adaptive immune system developed a strategy for monitoring the proteins synthesized in virtually any cell in the body, thereby preventing pathogens from 'hiding out' by adopting an intracellular lifestyle. To accomplish this, the immune system:
A) Co-opted the ubiquitin-proteasome system used by cell for protein turnover
B) Created a novel pathway using the immunoproteasome for generating peptides
C) Created a novel pathway to express foreign proteins on the cell surface
D) Took advantage of proteolytic enzymes present in endocytic vesicles
E) Engineered an immune-specific ubiquitin molecule for tagging foreign proteins
A) Co-opted the ubiquitin-proteasome system used by cell for protein turnover
B) Created a novel pathway using the immunoproteasome for generating peptides
C) Created a novel pathway to express foreign proteins on the cell surface
D) Took advantage of proteolytic enzymes present in endocytic vesicles
E) Engineered an immune-specific ubiquitin molecule for tagging foreign proteins

Unlock Deck
Unlock for access to all 30 flashcards in this deck.
Unlock Deck
k this deck
16
Empty MHC class I and MHC class II molecules are rapidly removed from the cell surface. This process prevents:
A) The accumulation of empty MHC molecules on the cell surface which would interfere with T cells recognizing pathogen-derived peptide:MHC complexes
B) Pathogens from evading the immune response by inducing peptide release from cell surface MHC molecules
C) MHC class I molecules from being internalized into endosomes and binding endosome-derived peptides
D) HLA-DM from trafficking to the cell surface with MHC class II
E) Inappropriate T cell recognition of healthy cells that are not infected, nor have ingested a pathogen
A) The accumulation of empty MHC molecules on the cell surface which would interfere with T cells recognizing pathogen-derived peptide:MHC complexes
B) Pathogens from evading the immune response by inducing peptide release from cell surface MHC molecules
C) MHC class I molecules from being internalized into endosomes and binding endosome-derived peptides
D) HLA-DM from trafficking to the cell surface with MHC class II
E) Inappropriate T cell recognition of healthy cells that are not infected, nor have ingested a pathogen

Unlock Deck
Unlock for access to all 30 flashcards in this deck.
Unlock Deck
k this deck
17
During MHC class I synthesis and folding in the endoplasmic reticulum (ER), a process of peptide editing takes place as the newly synthesized MHC class I protein is held in a 'peptide receptive' state by binding to the calreticulin:ERp57:tapasin complex. Peptide editing ensures that the MHC class I molecules that reach the cell surface have stable, high affinity binding for their peptide cargo. Peptide editing is important to the immune response because it:
A) Maintains high levels of surface MHC class I expression
B) Ensures that MHC class I molecules are not degraded in the ER
C) Retains the nascent MHC class I molecule in a peptide receptive state
D) Allows surface MHC class I molecules to bind new peptides from the extracellular milieu
E) Prevents surface MHC class I molecules from undergoing peptide exchange at the cell surface
A) Maintains high levels of surface MHC class I expression
B) Ensures that MHC class I molecules are not degraded in the ER
C) Retains the nascent MHC class I molecule in a peptide receptive state
D) Allows surface MHC class I molecules to bind new peptides from the extracellular milieu
E) Prevents surface MHC class I molecules from undergoing peptide exchange at the cell surface

Unlock Deck
Unlock for access to all 30 flashcards in this deck.
Unlock Deck
k this deck
18
A cell line carrying a mutation in a single gene is found to express very low levels of MHC class I on its surface. When infected with influenza virus, these cells are not recognized nor are they killed by a CD8 T cell line specific for an influenza peptide bound to the MHC class I protein expressed by these cells. Incubation of the mutant cell line with a large excess of this peptide in the cell culture medium overnight leads to the results shown in 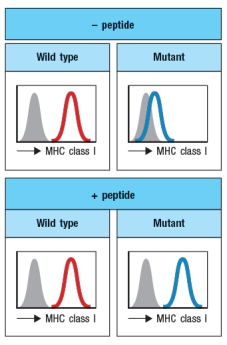
What is the most likely candidate for the gene that is defective in the mutant cell line?

What is the most likely candidate for the gene that is defective in the mutant cell line?

Unlock Deck
Unlock for access to all 30 flashcards in this deck.
Unlock Deck
k this deck
19
Peptide editing is an important component of antigen presentation for both MHC class I and MHC class II pathways, as it drives the preferential presentation of high-affinity binding peptides. For MHC class II peptide editing, HLA-DM plays a key role. In the absence of HLA-DM:
A) MHC class II molecules traffic to the cell surface with CLIP in their binding sites.
B) No MHC class II molecules are released to traffic to the cell surface.
C) MHC class II molecules bind to HLA-DO and are inhibited from binding peptides.
D) Pathogens can evade the immune system by blocking peptide exchange on MHC class II.
E) HLA-DO competes for high-affinity binding peptides with MHC class II molecules and blocks antigen presentation.
A) MHC class II molecules traffic to the cell surface with CLIP in their binding sites.
B) No MHC class II molecules are released to traffic to the cell surface.
C) MHC class II molecules bind to HLA-DO and are inhibited from binding peptides.
D) Pathogens can evade the immune system by blocking peptide exchange on MHC class II.
E) HLA-DO competes for high-affinity binding peptides with MHC class II molecules and blocks antigen presentation.

Unlock Deck
Unlock for access to all 30 flashcards in this deck.
Unlock Deck
k this deck
20
The genes encoding MHC proteins are closely linked with genes encoding proteins involved in antigen processing and presentation. This genetic linkage facilitates the coordinate regulation of these genes by interferons.

Unlock Deck
Unlock for access to all 30 flashcards in this deck.
Unlock Deck
k this deck
21
Alloreactivity refers to the ability of T cells to respond to allelic polymorphisms in MHC molecules when mixed with antigen-presenting cells from a genetically different individual. The T-cell receptors involved in alloreactive responses are recognizing amino acid sequences on foreign MHC molecules and do not interact at all with the peptides bound to these MHC molecules.

Unlock Deck
Unlock for access to all 30 flashcards in this deck.
Unlock Deck
k this deck
22
A family of six (mother, father, and four children) had two children with a history of chronic illness. Both children had repetitive infections of the sinuses, middle ears, and lungs due to a variety of respiratory viruses. Their other siblings were generally healthy and showed no signs of persistent or recurrent virus infections.
The two affected children had normal numbers of B cells, T cells, and NK cells in their blood. They also showed no defects in neutrophil function or in complement protein levels. The two children also had normal antibody levels to vaccine protein antigens, such as tetanus toxoid, and had normal T cell responses to antigens from the vaccine strain of Mycobacterium tuberculosis after being vaccinated.
Blood cells from one parent, one healthy child and the two affected children were examined for surface MHC protein expression by flow cytometry using two antibodies, one that recognizes all HLA class I proteins, and one that recognizes all HLA class II proteins. The results are shown in Figure
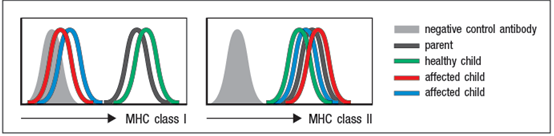
a) Analysis of HLA genotypes from the two affected children showed that they shared one haplotype of this locus. This haplotype encodes a common HLA-A allele, HLA-A2. Based on these data, is it likely that the two affected children have a point mutation (or mutations) in the coding sequence for HLA-A2? Why or why not?
b) Name two proteins that could be candidates for the defective gene in the two affected children.
To address which gene defect might be present in the affected children, peripheral blood cells were isolated from one healthy child and one affected child, and mRNA was isolated from the cells. Quantitative RT-PCR was performed to assess the levels of mRNA for the three HLA class I heavy chain genes (HLA-A, -B, and –C) and for the β2-microglobulin gene. The results are shown in Figure .
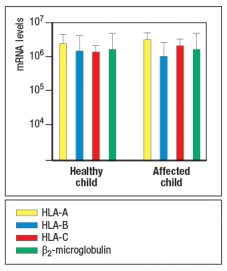
In a second experiment, Western blots were performed, confirming that cell lysates from both affected children contained normal levels of all three HLA class I heavy chain proteins and the β2-microglobulin protein.
c) Do these data eliminate any of your answers to part (b)?
In a final experiment, peripheral blood cells from the two affected children and one HLA-A2+ parent were transfected with a construct encoding the Hepatitis B virus surface antigen (HepBsAg), a protein that is currently used in the vaccine against Hepatitis B. This protein is normally synthesized and transported to the cell surface. In addition to the full length HepBsAg construct, cells were also transfected with a mini-gene, encoding just a single HLA-A2-binding peptide derived from Hepatitis B, amino acids 338–347. Using a cytolytic CD8 T cell clone specific for HepBsAg(338–347) peptide bound to HLA-A2, the transfected cells were tested for recognition by the CD8 T cell clone using an assay that measures target cell killing. Figure Q6.29C shows the results of this experiment.
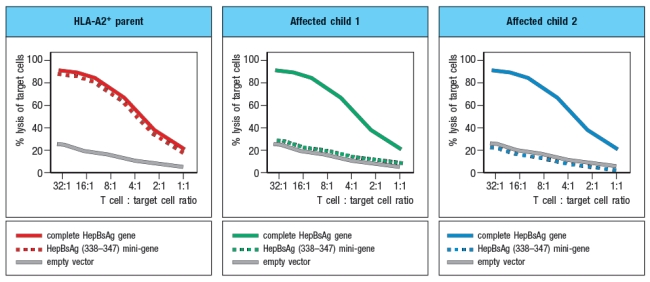
d) What is the most likely gene that is defective in the affected children?
The two affected children had normal numbers of B cells, T cells, and NK cells in their blood. They also showed no defects in neutrophil function or in complement protein levels. The two children also had normal antibody levels to vaccine protein antigens, such as tetanus toxoid, and had normal T cell responses to antigens from the vaccine strain of Mycobacterium tuberculosis after being vaccinated.
Blood cells from one parent, one healthy child and the two affected children were examined for surface MHC protein expression by flow cytometry using two antibodies, one that recognizes all HLA class I proteins, and one that recognizes all HLA class II proteins. The results are shown in Figure

a) Analysis of HLA genotypes from the two affected children showed that they shared one haplotype of this locus. This haplotype encodes a common HLA-A allele, HLA-A2. Based on these data, is it likely that the two affected children have a point mutation (or mutations) in the coding sequence for HLA-A2? Why or why not?
b) Name two proteins that could be candidates for the defective gene in the two affected children.
To address which gene defect might be present in the affected children, peripheral blood cells were isolated from one healthy child and one affected child, and mRNA was isolated from the cells. Quantitative RT-PCR was performed to assess the levels of mRNA for the three HLA class I heavy chain genes (HLA-A, -B, and –C) and for the β2-microglobulin gene. The results are shown in Figure .

In a second experiment, Western blots were performed, confirming that cell lysates from both affected children contained normal levels of all three HLA class I heavy chain proteins and the β2-microglobulin protein.
c) Do these data eliminate any of your answers to part (b)?
In a final experiment, peripheral blood cells from the two affected children and one HLA-A2+ parent were transfected with a construct encoding the Hepatitis B virus surface antigen (HepBsAg), a protein that is currently used in the vaccine against Hepatitis B. This protein is normally synthesized and transported to the cell surface. In addition to the full length HepBsAg construct, cells were also transfected with a mini-gene, encoding just a single HLA-A2-binding peptide derived from Hepatitis B, amino acids 338–347. Using a cytolytic CD8 T cell clone specific for HepBsAg(338–347) peptide bound to HLA-A2, the transfected cells were tested for recognition by the CD8 T cell clone using an assay that measures target cell killing. Figure Q6.29C shows the results of this experiment.

d) What is the most likely gene that is defective in the affected children?

Unlock Deck
Unlock for access to all 30 flashcards in this deck.
Unlock Deck
k this deck
23
Some CD1 molecules bind to glycosphingolipids, and are recognized by a subset of T cells known as invariant NKT (iNKT) cells. The ability of these T cells to recognize different glycolipid constituents from microorganisms when they are bound to CD1d places these cells in the 'innate immune' category. While iNKT cells do express a fully rearranged : T-cell receptor, one key feature of the T-cell receptors expressed on iNKT cells also places them in the 'innate immune' category. This feature is:
A) iNKT cells have a highly restricted T-cell receptor repertoire, with the majority of cells utilizing the same V and J rearrangement.
B) iNKT cells express receptors that are also expressed on NK cells.
C) iNKT cells express T-cell receptors that induce inhibitory, rather than activating signals.
D) iNKT cells do not generally express CD4 or CD8.
E) The T-cell receptors expressed on iNKT cells recognize both MHC class I and MHC class II molecules.
A) iNKT cells have a highly restricted T-cell receptor repertoire, with the majority of cells utilizing the same V and J rearrangement.
B) iNKT cells express receptors that are also expressed on NK cells.
C) iNKT cells express T-cell receptors that induce inhibitory, rather than activating signals.
D) iNKT cells do not generally express CD4 or CD8.
E) The T-cell receptors expressed on iNKT cells recognize both MHC class I and MHC class II molecules.

Unlock Deck
Unlock for access to all 30 flashcards in this deck.
Unlock Deck
k this deck
24
The extensive polymorphism of MHC genes in the population is thought to represent an evolutionary response to outflank the evasive strategies of pathogens. This polymorphism makes it difficult for pathogens to eliminate all potential MHC binding epitopes from their proteins. Based on this reasoning, it would seem advantageous for each individual to encode more than three different MHC class I and three different MHC class II genes per chromosome copy. If some individuals in the population had MHC loci that encoded 10 different MHC class I and 10 different MHC class II genes, the T cell repertoire in those individuals would likely be:
A) Much more diverse than in the rest of the individuals of that population
B) Much better at recognizing rare pathogens not encountered by most individuals in that population
C) Much less diverse than the rest of the individuals in that population
D) Much more alloreactive than the T cells found in the other individuals of that population
E) Very reactive to bacterial and viral superantigens
A) Much more diverse than in the rest of the individuals of that population
B) Much better at recognizing rare pathogens not encountered by most individuals in that population
C) Much less diverse than the rest of the individuals in that population
D) Much more alloreactive than the T cells found in the other individuals of that population
E) Very reactive to bacterial and viral superantigens

Unlock Deck
Unlock for access to all 30 flashcards in this deck.
Unlock Deck
k this deck
25
In the 1980s, a mutant strain of mice was identified, carrying amino acid changes in the MHC class II gene. This mutant strain was derived from C57Bl/6 mice, which carry the H-2b haplotype. Inbred H-2b mice express only one MHC class II protein, called Ab. The mutant strain, called 'bm12' was found to have 3 amino acid changes in the Ab protein, at positions 67, 70, and 71 of the Aβ chain. The positions of these amino acid changes on the MHC class II structure are shown below by the red circles in Figure . On the right, the side view diagram of MHC class II shows the direction of these three amino acid side chains. 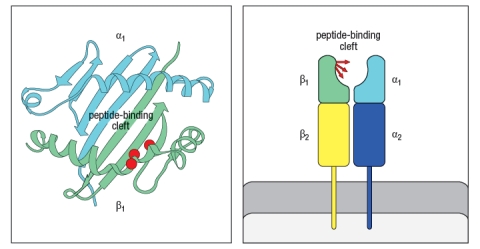
Initial experiments with wild-type C57Bl/6 mice and bm12 mice showed that the wild-type mice made a robust CD4 T cell response after immunization with the insulin protein isolated from a cow; in contrast, the bm12 mice failed to make any detectable response to this foreign protein. Epitope mapping studies identified amino acid residues 1–14 of the bovine insulin A chain as the peptide recognized by CD4 T cells from wild-type mice.
a) What is the most likely explanation for the failure of bm12 mice to make a CD4 T cell response to bovine insulin?
In a second set of experiments, T cells from wild-type (WT) or bm12 mice were mixed in vitro with antigen-presenting cells (APCs), in the presence or absence of the superantigen staphylococcal enterotoxin B (SEB), and T cell proliferation was measured. The data from these experiments are shown in Figure.
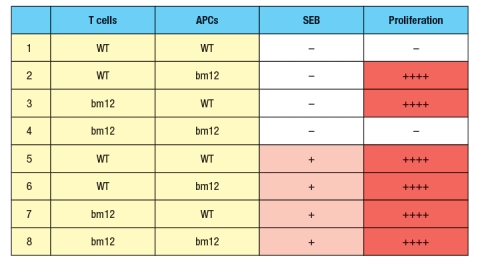
b) What is the explanation for the results in Rows 1–4 of the table?
c) Why does the T cell response to SEB (Rows 5–8) show a different pattern than the response to bovine insulin?
d) In the table above, T cell proliferation was measured after 4 days of incubation of T cells, APCs, +/- SEB. If one isolated the T cells at the end of the incubation for the six conditions in which robust proliferation was seen (Rows 2, 3, 5-8), and stained the T cells with each antibody (separately) from a panel of antibodies that recognize each of the mouse V domains (i.e., an antibody to V 1, an antibody to V 2, etc), what result would be expected?

Initial experiments with wild-type C57Bl/6 mice and bm12 mice showed that the wild-type mice made a robust CD4 T cell response after immunization with the insulin protein isolated from a cow; in contrast, the bm12 mice failed to make any detectable response to this foreign protein. Epitope mapping studies identified amino acid residues 1–14 of the bovine insulin A chain as the peptide recognized by CD4 T cells from wild-type mice.
a) What is the most likely explanation for the failure of bm12 mice to make a CD4 T cell response to bovine insulin?
In a second set of experiments, T cells from wild-type (WT) or bm12 mice were mixed in vitro with antigen-presenting cells (APCs), in the presence or absence of the superantigen staphylococcal enterotoxin B (SEB), and T cell proliferation was measured. The data from these experiments are shown in Figure.

b) What is the explanation for the results in Rows 1–4 of the table?
c) Why does the T cell response to SEB (Rows 5–8) show a different pattern than the response to bovine insulin?
d) In the table above, T cell proliferation was measured after 4 days of incubation of T cells, APCs, +/- SEB. If one isolated the T cells at the end of the incubation for the six conditions in which robust proliferation was seen (Rows 2, 3, 5-8), and stained the T cells with each antibody (separately) from a panel of antibodies that recognize each of the mouse V domains (i.e., an antibody to V 1, an antibody to V 2, etc), what result would be expected?

Unlock Deck
Unlock for access to all 30 flashcards in this deck.
Unlock Deck
k this deck
26
NKG2D is an activating receptor expressed on NK cells, : T cells, and some cytotoxic : T cells. When stressed or infected cells up-regulate receptors that bind to and activate NKG2D molecules, the stressed or infected cells will be killed. This pathway relies on the fact that stressed or infected cells up-regulate:
A) All classical MHC class I molecules
B) HLA-C molecules that bind KIRs
C) MHC class Ib genes such as MICA, MICB, and RAET1
D) Qa-1 and HLA-E molecules that bind leader peptides of other HLA class I molecules
E) HLA-G molecules just like those expressed on the fetal-derived cells in the placenta
A) All classical MHC class I molecules
B) HLA-C molecules that bind KIRs
C) MHC class Ib genes such as MICA, MICB, and RAET1
D) Qa-1 and HLA-E molecules that bind leader peptides of other HLA class I molecules
E) HLA-G molecules just like those expressed on the fetal-derived cells in the placenta

Unlock Deck
Unlock for access to all 30 flashcards in this deck.
Unlock Deck
k this deck
27
The MHC locus encodes a large number of genes spanning over four million bp of DNA. These include many genes involved in antigen processing and presentation, as well as receptors recognized by non-conventional T cells and natural killer (NK) cells. In addition, the MHC locus encodes genes with no function in immunity at all.

Unlock Deck
Unlock for access to all 30 flashcards in this deck.
Unlock Deck
k this deck
28
NKT cells that recognize microbial glycolipids bound to CD1 molecules comprise a class of T cells that shares features of both innate and adaptive immune cells. A second class of such cells are MAIT cells, that recognize antigens bound to the MHC class Ib molecule, MR1. What is the class of PAMP recognized by MAIT cells?

Unlock Deck
Unlock for access to all 30 flashcards in this deck.
Unlock Deck
k this deck
29
T cells expressing : T-cell receptors have been found to recognize a diversity of ligands, including pathogen-derived proteins, self-peptides, and stress-induced molecules. This pattern of antigen recognition shows similarity to that of iNKT cells and MAIT cells, suggesting that : T cells:
A) Do not play an important role in immunity, but likely have a non-immune function
B) Share features of both innate and adaptive immune cells
C) Are only able to respond when the host is infected with a virus such as herpes simplex virus
D) Are involved in maintaining the integrity of endothelial cells in the host
E) Are most important in responses to tumor cells that show stress responses
A) Do not play an important role in immunity, but likely have a non-immune function
B) Share features of both innate and adaptive immune cells
C) Are only able to respond when the host is infected with a virus such as herpes simplex virus
D) Are involved in maintaining the integrity of endothelial cells in the host
E) Are most important in responses to tumor cells that show stress responses

Unlock Deck
Unlock for access to all 30 flashcards in this deck.
Unlock Deck
k this deck
30
Several types of pathogens encode proteins that function as superantigens, which activate massive numbers of T cells in an individual. One example is the staphylococcal enterotoxins that cause food poisoning. These superantigens are the exception to the general rule that T cells only recognize specific peptide:MHC complexes, because they:
A) Induce activation of any T cell whose T-cell receptor uses a particular Vβ region bound by that superantigen
B) Simultaneously stimulate all of the T-cell receptors on a given T cell
C) Cover up the peptide-binding site, preventing MHC molecules from binding peptides
D) Activate a large number of T cells that are specifically recognizing peptides derived from the superantigen protein
E) Induce anti-microbial cytokine production that aids the immune system in clearing the pathogen
A) Induce activation of any T cell whose T-cell receptor uses a particular Vβ region bound by that superantigen
B) Simultaneously stimulate all of the T-cell receptors on a given T cell
C) Cover up the peptide-binding site, preventing MHC molecules from binding peptides
D) Activate a large number of T cells that are specifically recognizing peptides derived from the superantigen protein
E) Induce anti-microbial cytokine production that aids the immune system in clearing the pathogen

Unlock Deck
Unlock for access to all 30 flashcards in this deck.
Unlock Deck
k this deck


