Multiple Choice
Morphologically, Species A is very similar to four other species, B-E. Yet the nucleotide sequence deep within an intron in a gene shared by all five of these eukaryotic species is quite different in Species A compared to that of the other four species when one studies the nucleotides present at each position.
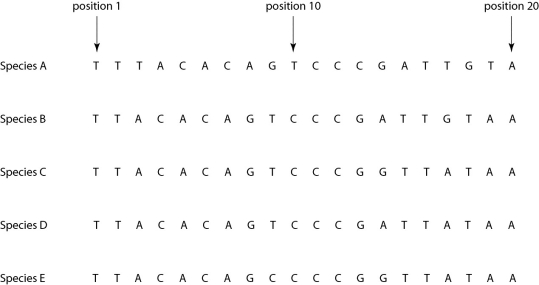 Figure 26.4
Figure 26.4
-If the sequence of Species A differs from that of the other four species due to simple misalignment, then what should the computer software find when it compares the sequence of Species A to those of the other four species?
A) The nucleotide at position 1 should be different in Species A, but the same in species B-E.
B) The nucleotide sequence of Species A should have long sequences that are nearly identical to those of the other species, but offset in terms of position number.
C) The sequences of species B-E, though different from that of Species A, should be identical to each other, without exception.
D) If the software compares, not nucleotide sequence, but rather the amino acid sequence of the actual protein product, then the amino acid sequences of species B-E should be similar to each other, but very different from that of Species A.
E) Computer software is useless in determining sequences of introns; it can only be used with exons.
Correct Answer:

Verified
Correct Answer:
Verified
Q2: The scientific discipline concerned with naming organisms
Q5: What is True of gene duplication (NOTE:
Q6: Use Figure 26.1 to answer the following
Q7: The following questions refer to the hypothetical
Q8: When it acts upon a gene, which
Q9: The following questions refer to the
Q10: The following questions refer to the hypothetical
Q11: Use Figure 26.1 to answer the following
Q25: The importance of computers and of computer
Q55: To apply parsimony to constructing a phylogenetic