Short Answer
You have used a variation of fluorescence in situ hybridization (FISH) called flow-FISH. In this technique, DNA probes are labeled with fluorescence and hybridized to their targets inside the cells in a tissue. The cells are then sorted using a fluorescence-activated cell sorter based on their fluorescent signal intensity. The probes in your experiment are designed to hybridize to human telomeric sequences at the ends of the chromosomes. You perform flow-FISH using these probes on two human cell types, an early primary fibroblast culture and a late secondary culture that is showing the signs of replicative cell senescence. The sorting results are presented in the following histograms. Which histogram (1 or 2) would you expect to correspond to the primary culture?
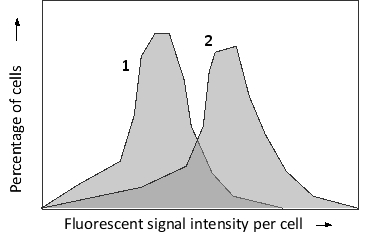
Correct Answer:

Verified
During replicative cell senes...View Answer
Unlock this answer now
Get Access to more Verified Answers free of charge
Correct Answer:
Verified
View Answer
Unlock this answer now
Get Access to more Verified Answers free of charge
Q56: Imagine a bistable system composed of two
Q57: Bacterial artificial chromosomes (BACs) …<br>A) are derived
Q58: The following schematic graph shows temperature change
Q59: Which of the following methods provides the
Q60: In the schematic graphs below, the red
Q62: In SDS-PAGE of proteins, …<br>A) the proteins
Q63: Which of the following can limit the
Q64: In a bacterium that is normally cylindrical
Q65: To create cellular factories for monoclonal antibody
Q66: The binding of protein A to two