Exam 10: Transcription in Bacteria
You have discovered a novel riboswitch that you think acts as a metabolite-responsive ribozyme. Design an experiment to demonstrate self-cleaving activity and metabolite specificity. Show sample positive results.
To demonstrate the self-cleaving activity and metabolite specificity of the novel riboswitch, we can design an experiment as follows:
1. In vitro transcription: First, we will synthesize the riboswitch sequence using in vitro transcription. This will allow us to obtain a pure sample of the riboswitch for our experiments.
2. Self-cleaving assay: Next, we will incubate the synthesized riboswitch with a buffer solution under different conditions. We will test for self-cleaving activity by adding a substrate that can be cleaved by the riboswitch. We will monitor the cleavage reaction using gel electrophoresis to visualize the cleaved products.
3. Metabolite specificity assay: To test for metabolite specificity, we will repeat the self-cleaving assay in the presence of different metabolites that the riboswitch is predicted to respond to. We will monitor the cleavage reaction in the presence of each metabolite and compare it to the reaction in the absence of any metabolite.
Sample positive results:
- In the self-cleaving assay, we observe a clear band corresponding to the cleaved product on the gel, indicating that the riboswitch is indeed capable of self-cleaving activity.
- In the metabolite specificity assay, we observe a significant increase in self-cleaving activity in the presence of a specific metabolite, while other metabolites have little to no effect on the cleavage reaction. This demonstrates the metabolite specificity of the riboswitch.
Overall, these results would provide strong evidence that the novel riboswitch acts as a metabolite-responsive ribozyme, exhibiting self-cleaving activity and specificity for a particular metabolite.
CAP is said to be responsible for positive regulation of the lac operon because:
D
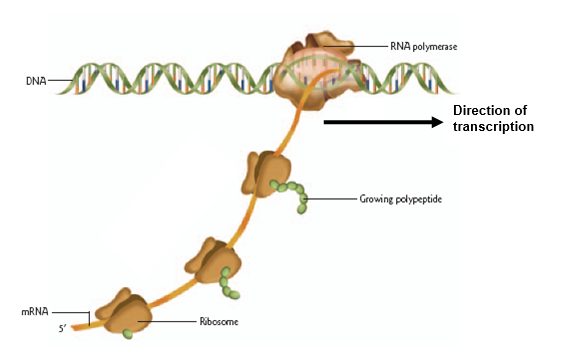
Draw a diagram of a prokaryotic gene being transcribed and translated. Show the nascent mRNA with ribosomes attached. With an arrow, indicate the direction of transcription.
Which part of a riboswitch-regulated mRNA binds to a regulatory metabolite molecule?
"Promoter clearance" refers to a step of transcription initiation in which
For bacterial operon induction, describe a specific example of the following: "The repressor and activators are DNA binding proteins that undergo allosteric modification."
A major difference between transcription in bacteria and eukaryotes is that
Consider E. coli cells, each having one of the following mutations:(a) A mutant lac operator sequence that cannot bind lac repressor(b) A mutant lac repressor that cannot bind to the lac operator(c) A mutant lac repressor that cannot bind to allolactose(d) A mutant lac promoter that cannot bind CAP plus cAMPWhat effect would each mutation have on the function of the lac operon (assuming no glucose is present)?
Draw a rough sketch of the structure of the bacterial RNA polymerase holoenzyme based on X-ray crystallography. Point out the position of the active site.
You are studying a new operon in bacteria involved in tyrosine biosynthesis.
(a) You sequence the operon and discover that it contains a short open reading frame at the 5′ end of the operon that contains two codons for tyrosine. What prediction would you make about this leader sequence, the RNA transcript, and the peptide that it encodes?
(b) How would you predict this operon is regulated; i.e., is it inducible or repressible by tyrosine? Why?
(c) Would this kind of regulation work in a eukaryotic cell? Why or why not?
Which statement best describes the role of ATP hydrolysis by the transcriptional terminator Rho?
Present a model to explain attenuation in the trp operon in E. coli. Is attenuation the main regulatory mechanism? Explain your answer.
Diagram a typical bacterial promoter. Exact sequences are not necessary.
You are studying a repressor protein that you suspect forms a DNA loop between two operator sites, one located very near the promoter and the other located at a distance site upstream. Describe an experiment to test your hypothesis.
A riboswitch is a domain within certain mRNAs that has all of the following characteristics, except:
Filters
- Essay(0)
- Multiple Choice(0)
- Short Answer(0)
- True False(0)
- Matching(0)