Short Answer
If two genomic polymorphic sites were randomly and independently associated with each other, one would expect the frequency of observing a combination of their allelic forms in an individual to be equal to the multiplication product of the probability of the alleles occurring separately. In many cases, however, this is not true. The degree of deviation from random association can be quantified and summarized in diagrams such as the one below. In this diagram, eight SNPs in a genomic region are shown and their pairwise deviation from random association is color-coded by different shades of red, such that the darkest red shade indicates the furthest deviation from random association. In contrast, those pairs that associate randomly are colored white. For example, SNPs A and F seem to associate randomly as indicated by the corresponding box marked with black borders. From this diagram, two haplotype blocks can be readily detected. What subset of the shown SNPs is not part of any of the two blocks? Write down the letter(s) as your answer, in alphabetical order, e.g. CEH. 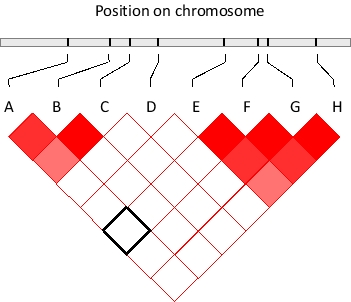
Correct Answer:

Verified
SNPs A, B, and C appear to co...View Answer
Unlock this answer now
Get Access to more Verified Answers free of charge
Correct Answer:
Verified
View Answer
Unlock this answer now
Get Access to more Verified Answers free of charge
Q43: Consider a promoter sequence with n binding
Q44: Consider a transcription regulatory protein (A) that
Q45: In sequence alignments such as those generated
Q46: Indicate whether ordinary agarose-gel electrophoresis (A), polyacrylamide-gel
Q47: You have used fluorescence-activated cell sorting to
Q49: You have discovered a mutant protein that
Q50: Consider the transcriptional circuits depicted in the
Q51: The quantitative output of a gene depends
Q52: Consider a transcription regulatory protein (A) that
Q53: You mated male and female heterozygous Oct4