Exam 4: DNA, Chromosomes, and Genomes
Exam 1: Cells and Genomes34 Questions
Exam 2: Cell Chemistry and Bioenergetics54 Questions
Exam 3: Proteins52 Questions
Exam 4: DNA, Chromosomes, and Genomes57 Questions
Exam 5: DNA Replication, Repair, and Recombination51 Questions
Exam 6: How Cells Read the Genome: From DNA to Protein58 Questions
Exam 7: Control of Gene Expression62 Questions
Exam 8: Analyzing Cells, Molecules, and Systems95 Questions
Exam 9: Visualizing Cells29 Questions
Exam 10: Membrane Structure26 Questions
Exam 11: Membrane Transport of Small Molecules and the Electrical Properties of Membranes46 Questions
Exam 12: Intracellular Compartments and Protein Sorting46 Questions
Exam 13: Intracellular Membrane Traffic54 Questions
Exam 14: Energy Conversion: Mitochondria and Chloroplasts49 Questions
Exam 15: Cell Signaling63 Questions
Exam 16: The Cytoskeleton75 Questions
Exam 17: The Cell Cycle57 Questions
Exam 18: Cell Death12 Questions
Exam 19: Cell Junctions and the Extracellular Matrix56 Questions
Exam 20: Cancer50 Questions
Exam 21: Development of Multicellular Organisms61 Questions
Exam 22: Stem Cells and Tissue Renewal45 Questions
Exam 23: Pathogens and Infection32 Questions
Exam 24: The Innate and Adaptive Immune Systems47 Questions
Select questions type
To study the chromatin remodeling complex SWR1, a researcher has prepared arrays of nucleosomes on long DNA strands that have been immobilized on magnetic beads. These nucleosomes are then incubated with an excess of the H2AZ-H2B dimer (which contains the histone variant H2AZ) in the presence or absence of SWR1 with or without ATP. She then separates the bead-bound nucleosomes (bound fraction) from the rest of the mix (unbound fraction) using a magnet, elutes the bound fraction from the beads, and performs SDS-PAGE on the samples. This is followed by a Western blot using an antibody specific to the H2AZ protein used in this experiment. The results are shown below, with the presence (+) or absence (-) of ATP, SWR1, or the H2AZ-H2B dimer in each incubation reaction indicated at the top of the corresponding lane. 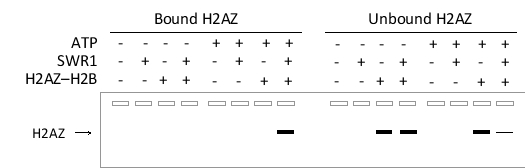 Which of the following statements is confirmed by the Western blot shown?
Which of the following statements is confirmed by the Western blot shown?
Free
(Multiple Choice)
4.9/5  (37)
(37)
Correct Answer:
A
Findings from a number of experiments on human chromatin have suggested that the DNA in our chromosomes is organized into loops of various lengths. Approximately how long is a typical loop (in nucleotide pairs of DNA)?
Free
(Multiple Choice)
4.8/5  (35)
(35)
Correct Answer:
C
Indicate whether each of the following descriptions better matches the major histones (M) or the histone variants (V). Your answer would be a six-letter string composed of letters M and V only, e.g. VVMVMV.
( ) They are more highly conserved over long evolutionary time scales.
( ) They are present in much smaller amounts in the cell.
( ) They are synthesized primarily during the S phase of the cell cycle.
( ) Their incorporation often requires histone exchange.
( ) They are often inserted into already-formed chromatin.
( ) They are assembled into nucleosomes just behind the replication fork.
Free
(Short Answer)
4.8/5  (29)
(29)
Correct Answer:
M
V
M
V
V
M
Nucleosomes that are positioned like beads on a string over a region of DNA can interact to form higher orders of chromatin structure. Which of the following factors can contribute to the formation of the 30-nm chromatin fiber from these nucleosomes?
(Multiple Choice)
4.9/5  (35)
(35)
Assume two isolated human communities with 500 individuals in each. If the same neutral mutation happens at the same time in two individuals, one from each community, what is the probability that it will be eventually fixed in both of the populations? How would the result change if the two communities fully interbreed? Write down the numbers in scientific notation and separate the two answers with a comma, e.g. 10-?, 3 × 10-².
Answers
(Short Answer)
4.8/5  (34)
(34)
You have performed a chromosome conformation capture (3C) experiment to study chromatin looping at a mouse gene cluster that contains genes A, B, and C, as well as a regulatory region R. In this experiment, you performed in situ chemical cross-linking of chromatin, followed by cleavage of DNA in the nuclear extract with a restriction enzyme, intramolecular ligation, and cross-link removal. Finally, a polymerase chain reaction (PCR) was carried out using a forward primer that hybridizes to a region in the active B gene, and one of several reverse primers, each of which hybridizes to a different location in the locus. The amounts of the PCR products were quantified and normalized to represent the relative cross-linking efficiency in each analyzed sample. You have plotted the results in the graph below. The same experiment has been done on two tissue samples: fetal liver (represented by red lines) and fetal muscle (blue lines).
Interaction of the A, B, and C genes with the regulatory R region is known to enhance expression of these genes. Indicate true (T) and false (F) statements below based on the results. Your answer would be a four-letter string composed of letters T and F only, e.g. TTTF.
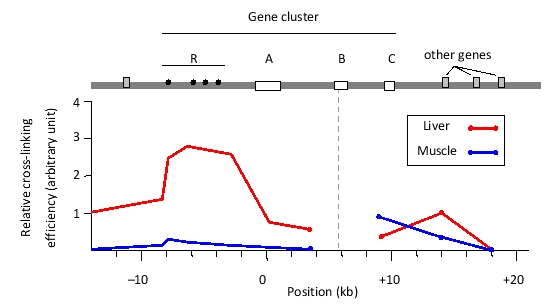 ( ) The B gene would be predicted to have higher expression in the fetal liver compared to the fetal muscle tissue.
( ) Interaction between the R region and the B gene involves the A gene looping out.
( ) Interaction between the R region and the B gene involves the C gene looping out.
( ) In fetal muscle, the B gene definitely does not engage in looping interactions with any other elements in the cluster.
( ) The B gene would be predicted to have higher expression in the fetal liver compared to the fetal muscle tissue.
( ) Interaction between the R region and the B gene involves the A gene looping out.
( ) Interaction between the R region and the B gene involves the C gene looping out.
( ) In fetal muscle, the B gene definitely does not engage in looping interactions with any other elements in the cluster.
(Short Answer)
4.7/5  (36)
(36)
Chromosome 3 contains nearly 200 million nucleotide pairs of our genome. If this DNA molecule could be laid end to end, how long would it be? The distance between neighboring base pairs in DNA is typically around 0.34 nm.
(Multiple Choice)
4.8/5  (33)
(33)
Indicate which numbered feature (1 to 5) in the schematic drawing below of the DNA double helix corresponds to each of the following. Your answer would be a five-digit number composed of digits 1 to 5 only, e.g. 52431.
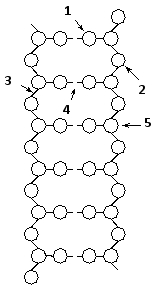 ( ) Hydrogen-bonding
( ) Covalent linkage
( ) Phosphate group
( ) Nitrogen-containing base
( ) Deoxyribose sugar
( ) Hydrogen-bonding
( ) Covalent linkage
( ) Phosphate group
( ) Nitrogen-containing base
( ) Deoxyribose sugar
(Short Answer)
4.8/5  (36)
(36)
Indicate which feature (1 to 4) in the schematic drawing below of a chromatin fiber corresponds to each of the following. Your answer would be a four-digit number composed of digits 1 to 4 only, e.g. 2431.
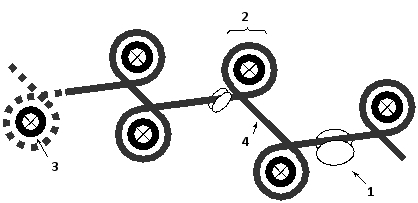 ( ) Nucleosome core particle
( ) Linker DNA
( ) Histone octamer
( ) Non-histone protein
( ) Nucleosome core particle
( ) Linker DNA
( ) Histone octamer
( ) Non-histone protein
(Short Answer)
4.9/5  (26)
(26)
Indicate whether each of the following histone modifications adds a negative charge to the histone (A), removes a positive charge from the histone (B), or does neither of these (C). Your answer would be a four-letter string composed of letters A, B, and C only, e.g. CABA.
( ) H3 lysine 9 acetylation
( ) H3 serine 10 phosphorylation
( ) H3 lysine 4 trimethylation
( ) H3 lysine 9 trimethylation
(Short Answer)
4.9/5  (33)
(33)
The two chromosomes in each of the 22 homologous pairs in our cells ...
(Multiple Choice)
4.8/5  (28)
(28)
In each of the following comparisons, indicate whether the molecular clock is expected to tick faster on average in the first (1) or the second (2) case. Your answer would be a four-digit number composed of digits 1 and 2 only, e.g. 2222.
( ) 1: The exons, or 2: the introns of a gene
( ) 1: The mitochondrial, or 2: the nuclear DNA of vertebrates
( ) 1: The first, or 2: the third position in synonymous codons
( ) 1: A gene, or 2: its pseudogene counterpart
(Short Answer)
4.9/5  (35)
(35)
Complete the DNA sequence below such that the final sequence is identical to that of the complementary strand. Your answer would be a seven-letter string composed of letters A, C, T, and G only, e.g. TTCTCAG.

(Short Answer)
4.8/5  (36)
(36)
Indicate true (T) and false (F) statements below regarding a genome and its evolution. Your answer would be a four-letter string composed of letters T and F only, e.g. FFTF.
( ) The genome of the last common ancestor of mammals can be investigated only if a DNA sample of the ancestor can be obtained.
( ) All of the "ultraconserved" elements found in the human genome have been shown to encode long noncoding RNAs.
( ) If a mouse carrying a homozygous deletion of a highly conserved genomic sequence survives and shows no noticeable defect, the highly conserved sequence has to be functionally unimportant.
( ) The "human accelerated regions" are genomic regions that are found in humans with no homologs in chimpanzees or other animals.
(Short Answer)
4.9/5  (43)
(43)
The regions of synteny between the chromosomes of two species can be visualized in dot plots. In the example shown in the following graph, a chromosome of a hypothetical species A has been aligned with the related chromosome in species B. Each dot in the plot represents the observation of high sequence identity between the two aligned chromosomes in a window located at the two corresponding chromosome positions. A series of close dots can make a continuous line. Choosing a sufficiently large window size allows a "clean" dot plot with solid lines that show only the long stretches of identity, allowing ancient large-scale rearrangements to be identified. Several chromosomal events can be detected in such dot plots. Indicate which feature (a to g) in the dot plot is best explained by each of the following events. Your answer would be a seven-letter string composed of letters a to g only, e.g. cdbagef. Each letter should be used only once.
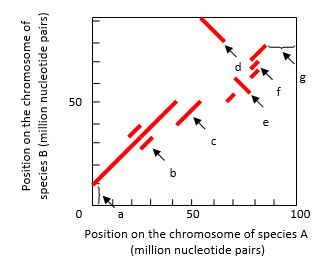 ( ) A duplication that exists in both species
( ) A duplication in species A only
( ) A triplication in species B only
( ) An inversion without relocation
( ) An inversion combined with relocation
( ) A deletion in species A
( ) A translocation in species A from a different chromosome
( ) A duplication that exists in both species
( ) A duplication in species A only
( ) A triplication in species B only
( ) An inversion without relocation
( ) An inversion combined with relocation
( ) A deletion in species A
( ) A translocation in species A from a different chromosome
(Short Answer)
4.9/5  (41)
(41)
Phylogenetic trees based on nucleotide or amino acid sequences can be constructed using various algorithms. One simple algorithm is based on a matrix of pairwise genetic distances (divergences) calculated after multiple alignment of the sequences. Imagine you have aligned a particular gene from different hominids (humans and the great apes), and have estimated the normalized number of nucleotide substitutions that have occurred in this gene in each pair of organisms since their divergence from their last common ancestor. You have obtained the following distance matrix.
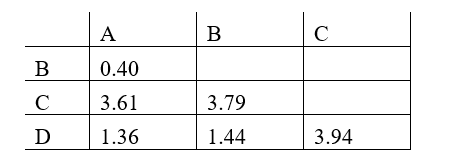 Answer the following question(s) based on this matrix.
-The following tree can be constructed from these distances assuming a constant molecular clock, meaning that the length of each horizontal branch corresponds to evolutionary time as well as to the relative genetic distance from the common ancestor that gave rise to that branch. Indicate which one of the species in the matrix (B to D) corresponds to branches 1 to 3, respectively. Your answer would be a three-letter string composed of letters B, C, and D only, e.g. DCB.
Answer the following question(s) based on this matrix.
-The following tree can be constructed from these distances assuming a constant molecular clock, meaning that the length of each horizontal branch corresponds to evolutionary time as well as to the relative genetic distance from the common ancestor that gave rise to that branch. Indicate which one of the species in the matrix (B to D) corresponds to branches 1 to 3, respectively. Your answer would be a three-letter string composed of letters B, C, and D only, e.g. DCB.
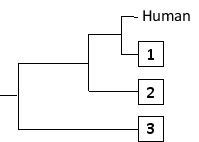
(Short Answer)
4.8/5  (32)
(32)
What are the consequences for the cell of the "fractal globule" arrangement of chromosome segments?
(Multiple Choice)
4.8/5  (32)
(32)
For the Human Genome Project, cloning of large segments of our genome was first made possible by the development of yeast artificial chromosomes, which are capable of propagating in the yeast Saccharomyces cerevisiae just like any of the organism's 16 natural chromosomes. In addition to the cloned human DNA, these artificial vectors were made to contain three elements that are necessary for them to function as a chromosome. What are these elements? Write down the names of the elements in alphabetical order, and separate them with commas, e.g. gene, histone, nucleosome.
(Short Answer)
4.8/5  (27)
(27)
A gene that had been turned off in a liver cell has just been induced to be highly expressed as the cell responds to a new metabolic load. What observations do you expect to accompany this change?
(Multiple Choice)
4.9/5  (34)
(34)
Showing 1 - 20 of 57
Filters
- Essay(0)
- Multiple Choice(0)
- Short Answer(0)
- True False(0)
- Matching(0)