Exam 7: Control of Gene Expression
Exam 1: Cells and Genomes34 Questions
Exam 2: Cell Chemistry and Bioenergetics54 Questions
Exam 3: Proteins52 Questions
Exam 4: DNA, Chromosomes, and Genomes57 Questions
Exam 5: DNA Replication, Repair, and Recombination51 Questions
Exam 6: How Cells Read the Genome: From DNA to Protein58 Questions
Exam 7: Control of Gene Expression62 Questions
Exam 8: Analyzing Cells, Molecules, and Systems95 Questions
Exam 9: Visualizing Cells29 Questions
Exam 10: Membrane Structure26 Questions
Exam 11: Membrane Transport of Small Molecules and the Electrical Properties of Membranes46 Questions
Exam 12: Intracellular Compartments and Protein Sorting46 Questions
Exam 13: Intracellular Membrane Traffic54 Questions
Exam 14: Energy Conversion: Mitochondria and Chloroplasts49 Questions
Exam 15: Cell Signaling63 Questions
Exam 16: The Cytoskeleton75 Questions
Exam 17: The Cell Cycle57 Questions
Exam 18: Cell Death12 Questions
Exam 19: Cell Junctions and the Extracellular Matrix56 Questions
Exam 20: Cancer50 Questions
Exam 21: Development of Multicellular Organisms61 Questions
Exam 22: Stem Cells and Tissue Renewal45 Questions
Exam 23: Pathogens and Infection32 Questions
Exam 24: The Innate and Adaptive Immune Systems47 Questions
Select questions type
In the following pedigree, females and males are indicated by circles and squares, respectively, and the presence of a rare disease caused by a loss-of-function mutation in an imprinted autosomal gene is indicated by black color. Is the gene maternally imprinted (M; i.e. not expressed from the maternally inherited chromosome) or paternally imprinted (P)? Write down M or P as your answer.
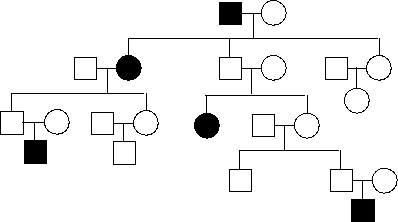
(Short Answer)
4.9/5  (34)
(34)
In the following schematic diagram of a region of a mammalian genome, genes A to D (triangles) are located in between a number of insulator elements (white squares) and barrier sequences (black squares). If the cis-regulatory sequence (oval) is bound by an abundant repressor protein, which gene would you expect to be expressed at a higher rate in this cell? 
(Short Answer)
4.8/5  (41)
(41)
Indicate whether each of the following descriptions better matches miRNAs (M) or siRNAs (S) in animal cells. Your answer would be a four-letter string composed of letters M and S only, e.g. MMMM.
( ) They usually cleave their target RNAs using the slicing activity of the Argonautes.
( ) They typically show perfect complementarity with their target RNA.
( ) They can bind to the RITS as well as the RISC complex.
( ) They seem to be the more ancient form of small noncoding RNAs.
(Short Answer)
4.8/5  (30)
(30)
In the following schematic diagram of five pairs of alternative RNA splicing patterns, which pair depicts exon skipping only? In exon skipping, an exon is spliced out of the RNA transcript along with its flanking introns. In the diagram, exons are colored dark blue and introns are colored yellow. Light blue indicates possible exons.
(Multiple Choice)
4.9/5  (29)
(29)
Indicate true (T) and false (F) statements below regarding DNA methylation in humans. Your answer would be a four-letter string composed of letters T and F only, e.g. TTTF.
( ) Methylation of adenines is the most common DNA methylation in humans.
( ) Methylated cytosine can be accidentally deaminated to produce thymine, leading to a C-to-T transition.
( ) Cytosine methylation often occurs within a 5?-CG-3? sequence.
( ) Shortly after fertilization, a genome-wide wave of demethylation takes place.
(Short Answer)
4.8/5  (38)
(38)
A certain region of a mammalian genome is transcribed at low but sustained levels using two flanking promoters. The RNA products are annealed together, processed, and used to recruit the RITS complex to this region, as well as chromatin modifying enzymes, RNA-dependent RNA polymerase, and a Dicer enzyme. Indicate whether this pathway is more closely related to that seen in miRNA (M), piRNA (P), or siRNA (S) interference pathways. Write down M, P, or S as your answer.
(Short Answer)
5.0/5  (32)
(32)
The following schematic drawing represents the activation of transcription for a eukaryotic gene (gene X). Indicate what component (A to D) in the drawing corresponds to each of the following. Your answer would be a four-letter string composed of letters A to D only, e.g. ADBC.
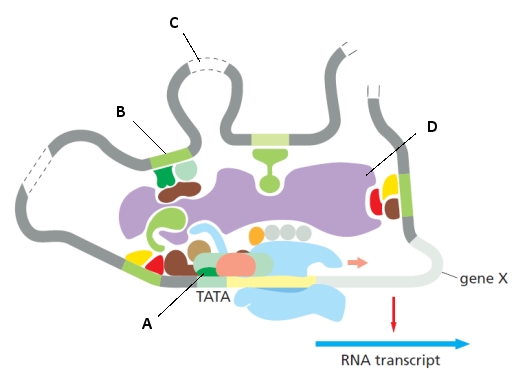 ( ) Mediator
( ) cis-Regulatory sequence
( ) A general transcription factor
( ) "Spacer" DNA that may encode lncRNAs
( ) Mediator
( ) cis-Regulatory sequence
( ) A general transcription factor
( ) "Spacer" DNA that may encode lncRNAs
(Short Answer)
4.9/5  (44)
(44)
Two transcription activators cooperate to recruit a coactivator to a cis-regulatory sequence and activate transcription of a nearby gene. If each of the activators increases the affinity of the coactivator for the reaction site (and therefore the rate of transcription) by 100-fold, how much would you expect the affinity to increase when both activators are bound to DNA compared to when none is bound?
(Multiple Choice)
4.9/5  (43)
(43)
Expression of the Even-skipped (Eve) gene in early Drosophila embryos is under the control of several transcription regulators. In one example, one of the Eve stripes is positioned near the anterior region of the embryo, and its regulatory module contains binding sites for Bicoid and Hunchback (activators) as well as Giant and Krüppel (inhibitors) such that the gene is expressed only in the region where concentrations of the two activators are high and the concentrations of the two inhibitors are low. A reporter gene can be placed under the control of this module, and it can be shown to form a stripe in the same place in the embryo as the corresponding stripe of Eve. Answer the following question(s) based on these findings.
-What would you expect to happen to the pattern of reporter expression in flies that lack the gene encoding Bicoid?
(Multiple Choice)
4.8/5  (40)
(40)
Beckwith-Wiedemann syndrome in humans is characterized by "overgrowth" in childhood, sometimes leading to unusually large parts of the body. An imprinted gene cluster on chromosome 11 is associated with this disease. The cluster contains several genes including Igf2 and H19. Igf2 encodes an insulin-like growth factor that is maternally imprinted, i.e. the maternal copy is not expressed. However, the DNA methylation pattern of this locus is not different between the two homologous chromosomes. On the other hand, H19 is also imprinted and its methylation pattern does differ between the two parental chromosomes. H19 is transcribed into a noncoding RNA that appears to silence the transcription of the Igf2 gene in cis. Would you expect the H19 locus to be hypermethylated in the maternally inherited chromosome (M) or paternally inherited chromosome (P)? Write down M or P as your answer.
(Short Answer)
4.9/5  (36)
(36)
To prevent spurious transcription from a gene, acetylation of histones-which is carried out by histone acetyl transferases ahead of a moving RNA polymerase II-is quickly reversed by histone deacetylases and histone methyl transferases in the wake of the polymerase, leaving a trail of specific methylated histones. Which of the following curves do you think better represents the distribution of this specific histone methylation mark with respect to a gene? 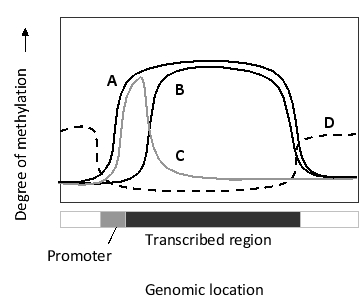
(Short Answer)
4.8/5  (44)
(44)
A DNA-binding protein recognizes a specific eight-nucelotide sequence in DNA. Assuming that its binding is perfectly specific, how many binding sites are expected to exist for it in human genomic DNA, which is composed of about 6 × 10⁹ nucleotide pairs? What about if the protein forms a homodimer? Note that the target sequence can be oriented either way in the double-stranded DNA.
(Multiple Choice)
4.8/5  (38)
(38)
In analysis using two-dimensional gel electrophoresis of the proteins expressed in different cell types, the number of spots that are different in different cells usually exceeds the number of common spots, and even the common spots can still have different intensities. The spots representing which of the following proteins would you expect to be among the common spots when compared across several cell types?
(Multiple Choice)
4.9/5  (38)
(38)
Indicate true (T) and false (F) statements below regarding riboswitches. Your answer would be a four-letter string composed of letters T and F only, e.g. TTTF.
( ) A riboswitch permits transcription elongation only when bound to its small-molecule ligand.
( ) Riboswitches appear to be a modern evolutionary invention.
( ) Riboswitches are often located near the 3? end of mRNAs, and therefore fold after the rest of the mRNA.
( ) Riboswitches are exclusively found in prokaryotes.
(Short Answer)
4.9/5  (35)
(35)
Comparing the bacterial CRISPR system and the eukaryotic RNAi mediated by siRNAs, indicate whether each of the following CRISPR components is most analogous to Argonaute (A), siRNA (S), or target mRNA (T) in the RNAi pathway. Your answer would be a three-letter string composed of letter A, S, and T only.
( ) Cas
( ) Viral DNA
( ) crRNA
(Short Answer)
4.9/5  (34)
(34)
In many animals, siRNAs can only function within the cell in which the siRNA is introduced. In contrast, the worm Caenorhabditis elegans can be fed with bacteria that synthesize double-stranded RNAs, and the RNAi spreads throughout the animal. A genetic screen to identify genes involved in systemic RNAi led to the discovery of Sid-1, a transmembrane protein expressed in most tissues in the adult worm. Loss-of-function mutations in the gene encoding Sid-1 restrict the RNAi activity to the cells surrounding the digestive tract after siRNA feeding. Ectopic expression of Sid-1 in Drosophila melanogaster cells that normally lack systemic RNAi and are unable to take up siRNAs from the medium, enables these cells to passively take up siRNA . These findings suggest that …
(Multiple Choice)
4.9/5  (44)
(44)
Imagine a gene encoding a pre-mRNA with twelve exons, ten of which can be alternatively spliced in vivo. Assuming that alternative splicing for this RNA only occurs through exon skipping, how many different proteins can possibly be made from this pre-mRNA as a result of alternative splicing?
(Multiple Choice)
4.9/5  (34)
(34)
The following sequence logo represents the preferred cis-regulatory sequences of an imaginary transcription regulator that functions as a dimer. Would you expect this sequence to be recognized by a homodimer (M) or a heterodimer (T)? Write down M or T as your answer.
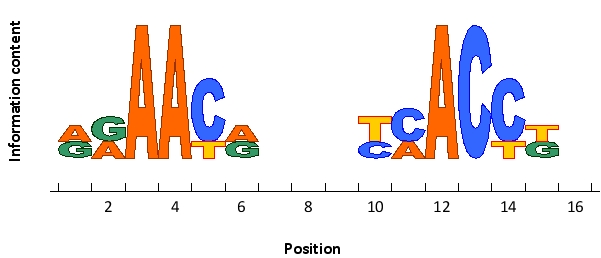
(True/False)
4.9/5  (40)
(40)
Expression of the Even-skipped (Eve) gene in early Drosophila embryos is under the control of several transcription regulators. In one example, one of the Eve stripes is positioned near the anterior region of the embryo, and its regulatory module contains binding sites for Bicoid and Hunchback (activators) as well as Giant and Krüppel (inhibitors) such that the gene is expressed only in the region where concentrations of the two activators are high and the concentrations of the two inhibitors are low. A reporter gene can be placed under the control of this module, and it can be shown to form a stripe in the same place in the embryo as the corresponding stripe of Eve. Answer the following question(s) based on these findings.
-What would you expect to happen to the pattern of reporter expression in flies that lack the gene encoding Giant?
(Multiple Choice)
4.8/5  (29)
(29)
Showing 21 - 40 of 62
Filters
- Essay(0)
- Multiple Choice(0)
- Short Answer(0)
- True False(0)
- Matching(0)