Exam 5: DNA and Chromosomes
The human genome is divided into linear segments and packaged into structures called chromosomes.What is the total number of chromosomes found in each of the somatic cells in your body?
D
For each of the following sentences, fill in the blanks with the best word or phrase selected from the list below.Not all words or phrases will be used; each word or phrase should be used only once.
bands extended kinetochore
chromatin homologous nonhomologous
chromosomes hybridization
condensation karyotype
In eukaryotic __________, DNA is complexed with proteins to form __________.The paternal and maternal copies of human Chromosome 1 are __________, whereas the paternal copy of Chromosome 1 and the maternal copy of Chromosome 3 are __________.Cytogeneticists can determine large-scale chromosomal abnormalities by looking at a patient's __________.Fluorescent molecules can be used to paint a chromosome, by a technique that employs DNA __________, and thereby to identify each chromosome by microscopy.
In eukaryotic chromosomes, DNA is complexed with proteins to form chromatin.The paternal and maternal copies of human Chromosome 1 are homologous, whereas the paternal copy of Chromosome 1 and the maternal copy of Chromosome 3 are nonhomologous.Cytogeneticists can determine large-scale chromosomal abnormalities by looking at a patient's karyotype.Fluorescent molecules can be used to paint a chromosome, by a technique that employs DNA hybridization, and thereby to identify each chromosome by microscopy.
The number of cells in an average-sized adult human is on the order of 1014.Use this information, and the estimate that the length of DNA contained in each cell is 2 m, to do the following calculations (look up the necessary distances and show your working):
A.Over how many miles would the total DNA from the average human stretch?
B.How many times would the total DNA from the average human wrap around the planet Earth at the Equator?
C.How many times would the total DNA from the average human stretch from Earth to the Sun and back?
D.How many times would the total DNA from the average human stretch from the Earth to Pluto and back?
A.2 × 1014 m = 124,274,238,447 miles.
B.The Earth's circumference at the Equator is 24,902 miles.The length of DNA from the average human body could wrap around the Earth 4,990,532 times.
C.The average distance from the Earth to the Sun is 93,000,000 miles.So, the round-trip distance is 186,000,000 miles.The length of DNA from the average human body could stretch from the Earth to the Sun and back 668 times.
D.The distance from the Earth to Pluto is, on average, about 39 × 93,000,000 miles.So, the round trip distance is 78 × 93,000,000 miles.The length of DNA from the average human body could stretch from the Earth to Pluto and back 17 times.
Several experiments were required to demonstrate how traits are inherited.Which scientist or team of scientists obtained definitive results demonstrating that DNA is the genetic molecule?
Use the terms listed to fill in the blanks in Figure 5-69.
A.A-T base pair
B.G-C base pair
C.deoxyribose
D.phosphodiester bonds
E.purine base
F.pyrimidine base 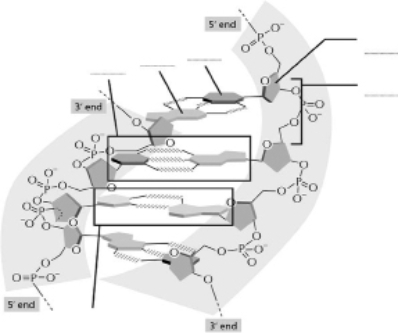 Figure 5-69
Figure 5-69
For each of the following sentences, fill in the blanks with the best word or phrase selected from the list below.Not all words or phrases will be used; each word or phrase should be used only once.
30-nm fiber heterochromatin linker active chromatin histone 1 loops axis histone 3 more beads-on-a-string histone 4 synaptic complex euchromatin less zigzag
Interphase chromosomes contain both darkly staining __________ and more lightly staining __________.Genes that are being transcribed are thought to be packaged in a __________ condensed type of euchromatin.Nucleosome core particles are separated from each other by stretches of __________ DNA.A string of nucleosomes coils up with the help of __________ to form the more compact structure of the __________.A __________ model describes the structure of the 30-nm fiber.The 30-nm chromatin fiber is further compacted by the formation of __________ that emanate from a central __________.
You are studying a newly identified chromatin-remodeling complex, which you call NICRC.You decide to run an in vitro experiment to characterize the activity of the purified complex.Your molecular toolbox includes: (1) a 400-base-pair DNA molecule that has a single recognition site for the restriction endonuclease EcoRI, an enzyme that cleaves internal sites on double-stranded DNA (dsDNA); (2) purified EcoRI enzyme; (3) purified DNase I, a DNA endonuclease that will cleave dsDNA at nonspecific sites if they are exposed; and (4) core octamer histones.You are able to assemble core nucleosomes on this DNA template and test for NICRC activity.Figure 5-67A illustrates the DNA template used and indicates both the location of the EcoRI cleavage site and the size of the DNA fragments that are produced when it cuts.Figure 5-67B illustrates how the DNA molecules in your experiment looked after separation according to size by using gel electrophoresis.Your experiment had a total of six samples, each of which was treated according to the legend below the gel.The sizes of the DNA fragments observed are indicated on the left side of the gel. (A)
 (B)
(B)
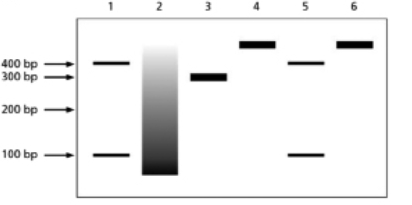 1. ECORI
2. DNase I
3. core octamer incubation, later treated with DNase I
4. core octamer incubation, later treated with EcoRI
5. core octamer incubation, addition of NICRC + ATP, later treated with EcoRI
6. core octamer incubation, addition of NICRC + ADP, later treated with EcoRI
Figure 5-67
A.Explain the results in lanes 1-4 and why it is important to have this information before you begin to test your remodeling complex.
B.What can you conclude about your purified remodeling complex from the results in lanes 5 and 6?
1. ECORI
2. DNase I
3. core octamer incubation, later treated with DNase I
4. core octamer incubation, later treated with EcoRI
5. core octamer incubation, addition of NICRC + ATP, later treated with EcoRI
6. core octamer incubation, addition of NICRC + ADP, later treated with EcoRI
Figure 5-67
A.Explain the results in lanes 1-4 and why it is important to have this information before you begin to test your remodeling complex.
B.What can you conclude about your purified remodeling complex from the results in lanes 5 and 6?
The classic "beads-on-a-string" structure is the most decondensed chromatin structure possible and is produced experimentally.Which chromatin components are NOT retained when this structure is generated?
Gene A, which is normally expressed, has been moved by DNA recombination near an area of heterochromatin.None of the daughter cells produced after this recombination event express gene A, even though its DNA sequence is unchanged.Explain this observation.
Stepwise condensation of linear DNA happens in five different packing processes.Which of the following four processes has a direct requirement for histone H1?
Fred Griffith studied two strains of Streptococcus pneumonia, one that causes a lethal infection when injected into mice, and a second that is harmless.He observed that pathogenic bacteria that have been killed by heating can no longer cause an infection.But when these heat-killed bacteria are mixed with live, harmless bacteria, this mixture is capable of infecting and killing a mouse.What did Griffith conclude from this experiment?
Origins of replication typically have a relatively high number of A-T base pairs.How does this sequence feature relate to the function of these DNA regions?
Most eukaryotic cells only express 20-30% of the genes they possess.The formation of heterochromatin maintains the other genes in a transcriptionally silent (unexpressed) state.Which histone modification is associated with the formation of the most common type of heterochromatin?
Which of the following DNA strands can form a DNA duplex by pairing with itself at each position?
Nucleosomes are formed when DNA wraps __________ times around the histone octamer in a __________ coil.
Mitotic chromosomes were first visualized with the use of very simple tools: a basic light microscope and some dyes.Which of the following characteristics of mitotic chromosomes reflects how they were named?
The N-terminal tail of histone H3 can be extensively modified, and depending on the number, location, and combination of these modifications, these changes may promote the formation of heterochromatin.What is the result of heterochromatin formation?
Filters
- Essay(0)
- Multiple Choice(0)
- Short Answer(0)
- True False(0)
- Matching(0)