Exam 20: Genomics and Proteomics
Exam 1: Introduction to Genetics65 Questions
Exam 2: Chromosomes and Cellular Reproduction62 Questions
Exam 3: Basic Principles of Heredity65 Questions
Exam 4: Sex Determination and Sex-Linked Characteristics87 Questions
Exam 5: Extensions and Modifications of Basic Principles93 Questions
Exam 6: Pedigree Analysis, Applications, and Genetic Testing78 Questions
Exam 7: Linkage, Recombination, and Eukaryotic Gene Mapping65 Questions
Exam 8: Chromosome Variation68 Questions
Exam 9: Bacterial and Viral Genetic Systems71 Questions
Exam 10: DNA: the Chemical Nature of the Gene82 Questions
Exam 11: Chromosome Structure and Organelle Dna83 Questions
Exam 12: DNA Replication and Recombination61 Questions
Exam 13: Transcription80 Questions
Exam 14: Rna Molecules and Rna Processing75 Questions
Exam 15: The Genetic Code and Translation76 Questions
Exam 16: Control of Gene Expression in Prokaryotes68 Questions
Exam 17: Control of Gene Expression in Eukaryotes64 Questions
Exam 18: Gene Mutations and Dna Repair100 Questions
Exam 19: Molecular Genetic Analysis and Biotechnology72 Questions
Exam 20: Genomics and Proteomics79 Questions
Exam 21: Epigenetics55 Questions
Exam 22: Developmental Genetics and Immunogenetics63 Questions
Exam 23: Cancer Genetics74 Questions
Exam 24: Quantitative Genetics81 Questions
Exam 25: Population Genetics69 Questions
Exam 26: Evolutionary Genetics63 Questions
Select questions type
Crossing over is often reduced around centromeric regions of chromosomes. If you were trying to construct a genetic map of two linked marker loci in this region, what result might you obtain?
Free
(Multiple Choice)
4.9/5  (36)
(36)
Correct Answer:
A
What information CANNOT be learned from the structure of a protein?
Free
(Multiple Choice)
4.9/5  (36)
(36)
Correct Answer:
E
The set of all proteins encoded by the genome is called the:
(Multiple Choice)
4.8/5  (42)
(42)
A set of overlapping DNA fragments that form a contiguous stretch of DNA is called a:
(Multiple Choice)
4.9/5  (35)
(35)
Use the following to answer questions
A researcher is interested in studying Drosophila genes that are turned on in response to UV light exposure. She uses a microarray to carry out her experiment and is particularly interested in eight genes shown in the microarray below.
Vertical stripes mean hybridization to UV light-exposed cell cDNA only.
Horizontal stripes mean hybridization to nonexposed cell cDNA only (control).
Completely filled in means hybridization to both cDNA types.
Blank means hybridization to neither cell's cDNA. 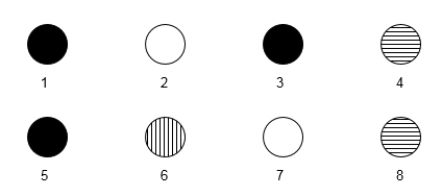 -Which genes appear to be upregulated in the UV light-exposed group?
-Which genes appear to be upregulated in the UV light-exposed group?
(Multiple Choice)
4.9/5  (31)
(31)
Use the following to answer questions
A researcher is interested in studying Drosophila genes that are turned on in response to UV light exposure. She uses a microarray to carry out her experiment and is particularly interested in eight genes shown in the microarray below.
Vertical stripes mean hybridization to UV light-exposed cell cDNA only.
Horizontal stripes mean hybridization to nonexposed cell cDNA only (control).
Completely filled in means hybridization to both cDNA types.
Blank means hybridization to neither cell's cDNA.  -Which genes appear to be involved in the UV light stress response in Drosophila?
-Which genes appear to be involved in the UV light stress response in Drosophila?
(Multiple Choice)
4.9/5  (35)
(35)
You have determined that a bacterial strain you are working with contains a single type of plasmid. You isolate the plasmid DNA and digest separate portions of it with each of two different restriction enzymes, BamHI and HpaI, and also perform a double digest using both enzymes. You then fractionate the enzyme digests on an agarose gel and stain the gel with ethidium bromide to visualize the restriction fragment patterns. Your results are shown below. Size markers (in nucleotide base pairs) are indicated at the left side of the gel. Using these data, construct a possible restriction map for the plasmid.
(Essay)
4.7/5  (39)
(39)
A human gene with a disease phenotype is going to be mapped by positional cloning. Which would be the MOST useful for this task?
(Multiple Choice)
4.9/5  (25)
(25)
In mass spectrometry, a molecule is ionized and its migration rate in an electrical field is determined. What information does the migration rate tell about the molecule?
(Multiple Choice)
4.9/5  (28)
(28)
Which class of genes is the result of an ancient gene duplication?
(Multiple Choice)
4.8/5  (37)
(37)
A new species of plant was discovered and its genome was sequenced. A BLAST search was performed on a small section of one of the chromosomes and one gene in this region showed 98% similarity to a gene in Arabidopsis. Based on this information, which of the following statements is FALSE?
(Multiple Choice)
4.9/5  (36)
(36)
Crossing over is often reduced around centromeric regions of chromosomes. If you were trying to construct a genetic map of two linked marker loci in this region, how would the genetic map correspond to the physical map?
(Multiple Choice)
4.7/5  (22)
(22)
Use the following to answer questions
The following microarray data are shown for four human genes. Two samples were hybridized to the microarray in this experiment. One sample was cDNA generated from mRNA of a tissue culture cell line that was originally derived from a B-cell leukemia. The other sample was cDNA generated from normal, mature neural tissue.
Vertical stripes mean hybridization to cancerous B-cell cDNA only.
Horizontal stripes mean hybridization to normal neural cell cDNA only.
Completely filled in means hybridization to both cDNA types.
Blank means hybridization to neither cell's cDNA. 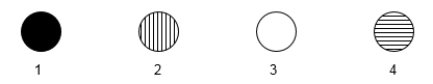 -One spot on the microarray contains sequences unique to a gene for an RNA polymerase subunit. One spot contains sequences unique to the skeletal muscle myosin gene. Which spot would be expected to contain the myosin gene sequences?
-One spot on the microarray contains sequences unique to a gene for an RNA polymerase subunit. One spot contains sequences unique to the skeletal muscle myosin gene. Which spot would be expected to contain the myosin gene sequences?
(Multiple Choice)
4.9/5  (35)
(35)
How can a BLAST search reveal information about protein function?
(Short Answer)
4.9/5  (23)
(23)
Explain why genetic and physical map distances may differ in relative distances between two genes on a chromosome.
(Essay)
4.9/5  (41)
(41)
The transcription factor Twist helps specify development of the mesoderm in the Drosophila embryo. You wish to identify other genes that Twist (as a transcription factor) switches on or off. You have wild-type embryos available, as well as embryos that are mutant for the Twist gene. Design an experiment using microarrays that could allow you to identify genes switched on or off by the Twist transcription factor.
(Essay)
4.8/5  (30)
(30)
You are attempting to generate a physical map of the bacteriophage genome using restriction enzyme digestions. You digest the phage DNA with EcoRI or NcoI or both, and observe the fragment sizes listed below. You know that bacteriophage has a linear, not circular, genome. EcoRI NcoI EcoRI/NcoI 21.2 19.3 19.3 7.4 16.4 7.4 5.8 4.6 5.1 5.7 4.2 3.9 4.9 4.0 3.5 3.5 2.7 2.2 1.9 1.8 0.7 a. How many times does the EcoRI enzyme cut the DNA? How many times does the NcoI enzyme cut the DNA?
b. Use the fragment size data to generate a physical map of the bacteriophage genome, indicating the order and relative positions of the restriction enzyme recognition sites.
(Essay)
4.7/5  (36)
(36)
Below is a genetic map for three loci. Which two of these loci would show the smallest physical distance in base pairs, if the frequency of crossing over is equal in this chromosome region? Explain your answer. A B C
(Essay)
4.9/5  (35)
(35)
Showing 1 - 20 of 79
Filters
- Essay(0)
- Multiple Choice(0)
- Short Answer(0)
- True False(0)
- Matching(0)