Exam 6: Mechanisms of Enzymes
Exam 1: Introduction to Biochemistry72 Questions
Exam 2: Water94 Questions
Exam 3: Amino Acids and the Primary Structures of Proteins107 Questions
Exam 4: Proteins: Three-Dimensional Structure and Function116 Questions
Exam 5: Properties of Enzymes91 Questions
Exam 6: Mechanisms of Enzymes88 Questions
Exam 7: Coenzymes and Vitamins93 Questions
Exam 8: Carbohydrates92 Questions
Exam 9: Lipids and Membranes95 Questions
Exam 10: Introduction to Metabolism87 Questions
Exam 11: Glycolysis88 Questions
Exam 12: Gluconeogenesis, the Pentose Phosphate Pathway, and Glycogen Metabolism90 Questions
Exam 13: The Citric Acid Cycle93 Questions
Exam 14: Electron Transport and Atp Synthesis95 Questions
Exam 15: Photosynthesis89 Questions
Exam 16: Lipid Metabolism89 Questions
Exam 17: Amino Acid Metabolism84 Questions
Exam 18: Nucleotide Metabolism81 Questions
Exam 19: Nucleic Acids95 Questions
Exam 20: DNA Replication, repair, and Recombination89 Questions
Exam 21: Transcription and RNA Processing91 Questions
Exam 22: Protein Synthesis99 Questions
Select questions type
Most Km values of enzymes for their substrates are on the order of ________ M.
(Multiple Choice)
4.8/5  (24)
(24)
Intermediates are more stable and have longer lifetimes than transition states.
(True/False)
5.0/5  (34)
(34)
The role of serine at the active site of serine proteases is to act as a(n)________ catalyst,while the histidine residue serves as a(n)________ catalyst.
(Multiple Choice)
4.8/5  (38)
(38)
Which will be different for a catalyzed reaction versus an uncatalyzed reaction?
(Multiple Choice)
4.8/5  (37)
(37)
The catalytic triad of chymotrypsin and other serine proteases consists of
(Multiple Choice)
4.8/5  (42)
(42)
The substrate specificity of serine proteases is primarily due to
(Multiple Choice)
4.8/5  (38)
(38)
An enzyme stabilizes the transition state that is bound in the active site.What effect will this have on the energy diagram below? The diagram shown below is for the uncatalyzed reaction. 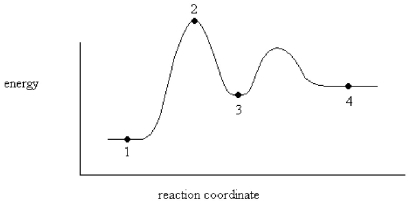
(Multiple Choice)
4.7/5  (32)
(32)
The reaction catalyzed by a certain phosphatase enzyme is found to follow ping-pong kinetics and involves the transfer of a phosphate group from substrate A to substrate B.Which mode of catalysis is likely for this reaction?
(Multiple Choice)
5.0/5  (33)
(33)
Transitions states bind to their enzymes more tightly than their substrates do.
(True/False)
4.9/5  (36)
(36)
What shape would a graph of reaction velocity versus pH have for an enzyme that uses both a proton donor and a proton acceptor during catalysis (both acid and base catalysis)?
(Multiple Choice)
4.8/5  (35)
(35)
Enzyme-catalyzed reactions that are diffusion-controlled reactions can never proceed any faster than the rate determined by random collision rates.
(True/False)
4.8/5  (33)
(33)
Reaction intermediates are impossible to isolate experimentally.
(True/False)
4.9/5  (36)
(36)
Superoxide dismutase and triosephosphate isomerase are two enzymes that catalyze diffusion-controlled reactions.
(True/False)
4.7/5  (42)
(42)
Radicals are stable,unreactive species that have an unpaired electron.
(True/False)
4.8/5  (36)
(36)
SOD (superoxide dismutase)is an enzyme that reacts faster than the rate of diffusion of the substrate to the active site due to
(Multiple Choice)
4.9/5  (27)
(27)
Glycoside hydrolases such as bacterial cellulase have been shown to differ in mechanisms from that of lysozyme.They
(Multiple Choice)
4.9/5  (30)
(30)
Transition state analogs are usually more potent inhibitors of enzyme activity than substrate analogs.
(True/False)
4.8/5  (32)
(32)
An enzyme's active site contains an arginine residue and a glutamate residue with pKa's of 2.9 and 9.1,respectively.Both residues are actively involved in the catalytic mechanism and they are the only two ionizable residues in the active site.What would you expect for the optimum pH of the enzyme?
(Multiple Choice)
5.0/5  (36)
(36)
The roles of amino acid residues at the active site of enzymes can be determined by removing certain residues using the technique of
(Multiple Choice)
4.8/5  (38)
(38)
Showing 61 - 80 of 88
Filters
- Essay(0)
- Multiple Choice(0)
- Short Answer(0)
- True False(0)
- Matching(0)