Exam 8: Control of Gene Expression
Exam 1: Cells: The Fundamental Units of Life64 Questions
Exam 2: Chemical Components of Cells74 Questions
Exam 3: Energy, Catalysis, and Biosynthesis73 Questions
Exam 4: Protein Structure and Function71 Questions
Exam 5: DNA and Chromosomes69 Questions
Exam 6: DNA Replication and Repair61 Questions
Exam 7: From DNA to Protein62 Questions
Exam 8: Control of Gene Expression68 Questions
Exam 9: How Genes and Genomes Evolve60 Questions
Exam 10: Analyzing the Structure and Function of Genes59 Questions
Exam 11: Membrane Structure57 Questions
Exam 12: Transport Across Cell Membranes67 Questions
Exam 13: How Cells Obtain Energy From Food71 Questions
Exam 14: Energy Generation in Mitochondria and Chloroplasts72 Questions
Exam 15: Intracellular Compartments and Protein Transport55 Questions
Exam 16: Cell Signaling60 Questions
Exam 17: Cytoskeleton59 Questions
Exam 18: The Cell-Division Cycle67 Questions
Exam 19: Sexual Reproduction and the Power of Genetics61 Questions
Exam 20: Cell Communities: Tissues, Stem Cells, and Cancer57 Questions
Select questions type
Label the following structures in Figure 8-51. 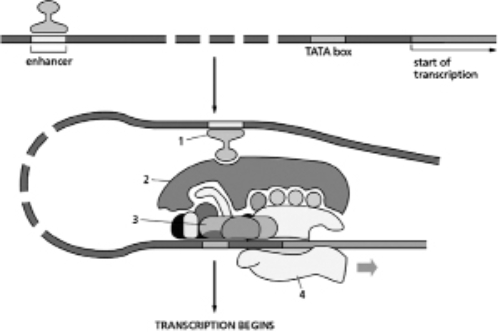 Figure 8-51
A.activator protein
B.RNA polymerase
C.general transcription factors
D.Mediator
Figure 8-51
A.activator protein
B.RNA polymerase
C.general transcription factors
D.Mediator
(Essay)
4.7/5  (38)
(38)
You are interested in understanding the gene regulation of Lkp1, a protein that is normally produced in liver and kidney cells in mice.Interestingly, you find that the LKP1 gene is not expressed in heart cells.You isolate the DNA upstream of the LKP1 gene, place it upstream of the gene for green fluorescent protein (GFP), and insert this entire piece of recombinant DNA into mice.You find GFP expressed in liver and kidney cells but not in heart cells, an expression pattern similar to the normal expression of the LKP1 gene.Further experiments demonstrate that there are three regions in the promoter, labeled A, B, and C in Figure 8-16, that contribute to this expression pattern.Assume that a single and unique transcription factor binds each site such that protein X binds site A, protein Y binds site B, and protein Z binds site C.You want to determine which region is responsible for tissue-specific expression, and create mutations in the promoter to determine the function of each of these regions.In Figure 8-16, if the site is missing, it is mutated such that it cannot bind its corresponding transcription factor.  Figure 8-16
Which of the following proteins is likely to act as a gene repressor?
Figure 8-16
Which of the following proteins is likely to act as a gene repressor?
(Multiple Choice)
4.9/5  (34)
(34)
Which of the following statements about iPS cells is FALSE?
(Multiple Choice)
4.8/5  (39)
(39)
Which of the following examples does NOT describe a mechanism of post-transcriptional control of gene expression?
(Multiple Choice)
4.8/5  (38)
(38)
Which of the following statements about the Lac operon is FALSE?
(Multiple Choice)
4.7/5  (39)
(39)
How are most eukaryotic transcription regulators able to affect transcription when their binding sites are far from the promoter?
(Multiple Choice)
4.8/5  (39)
(39)
You are interested in understanding the gene regulation of Lkp1, a protein that is normally produced in liver and kidney cells in mice.Interestingly, you find that the LKP1 gene is not expressed in heart cells.You isolate the DNA upstream of the LKP1 gene, place it upstream of the gene for green fluorescent protein (GFP), and insert this entire piece of recombinant DNA into mice.You find GFP expressed in liver and kidney cells but not in heart cells, an expression pattern similar to the normal expression of the LKP1 gene.Further experiments demonstrate that there are three regions in the promoter, labeled A, B, and C in Figure 8-16, that contribute to this expression pattern.Assume that a single and unique transcription factor binds each site such that protein X binds site A, protein Y binds site B, and protein Z binds site C.You want to determine which region is responsible for tissue-specific expression, and create mutations in the promoter to determine the function of each of these regions.In Figure 8-16, if the site is missing, it is mutated such that it cannot bind its corresponding transcription factor.  Figure 8-16
Experiment 1 in Figure 8-18 is the positive control, demonstrating that the region of DNA upstream of the gene for GFP results in a pattern of expression that we normally find for the LKP1 gene.Experiment 2 shows what happens when the sites for binding factors X, Y, and Z are removed.Which experiment above demonstrates that factor X alone is sufficient for expression of LPK1 in the kidney?
Figure 8-16
Experiment 1 in Figure 8-18 is the positive control, demonstrating that the region of DNA upstream of the gene for GFP results in a pattern of expression that we normally find for the LKP1 gene.Experiment 2 shows what happens when the sites for binding factors X, Y, and Z are removed.Which experiment above demonstrates that factor X alone is sufficient for expression of LPK1 in the kidney?
(Multiple Choice)
4.9/5  (37)
(37)
You are interested in studying the transcriptional regulation of the Gip1 promoter.The Gip1 promoter contains a binding site for the Jk8 protein that overlaps with the binding site for the Pa5 protein.Jk8 and Pa5 cannot bind DNA at the same time, but both proteins are present at high levels in adult liver cells.The binding sites for Jk8 and Pa5 are shown in Figure 8-24.  Figure 8-24
Jk8 binds to site A while Pa5 binds to site B.You create mutations that remove the nonoverlapping sequences of either binding site A or B, and examine Gip1 mRNA production in adult liver cells that contain these mutations.The data you obtain from these experiments are shown in Table 8-24. experiment number 1 2 3 4 binding site A B ++ +- -+ -- Gip1 mRNA levels high hight none none
Table 8-24
Which transcription regulatory protein is bound in experiment #1 (the normal situation)? Explain.
Figure 8-24
Jk8 binds to site A while Pa5 binds to site B.You create mutations that remove the nonoverlapping sequences of either binding site A or B, and examine Gip1 mRNA production in adult liver cells that contain these mutations.The data you obtain from these experiments are shown in Table 8-24. experiment number 1 2 3 4 binding site A B ++ +- -+ -- Gip1 mRNA levels high hight none none
Table 8-24
Which transcription regulatory protein is bound in experiment #1 (the normal situation)? Explain.
(Essay)
4.8/5  (33)
(33)
You are studying a set of mouse genes whose expression increases when cells are exposed to the hormone cortisol, and you believe that the same cortisol-responsive transcriptional activator regulates all of these genes.If your hypothesis is correct, which of the following statements below should be TRUE?
(Multiple Choice)
4.9/5  (33)
(33)
Which of the following statements about transcriptional regulators is FALSE?
(Multiple Choice)
4.9/5  (36)
(36)
Fill in the blanks with the best word or phrase selected from the list below.Not all words or phrases will be used; each word or phrase should be used only once.
allosteric negatively positively constitutively operator promoter induced operon repressed
The genes of a bacterial __________ are transcribed into a single mRNA.Many bacterial promoters contain a region known as a/an __________, to which a specific transcription regulator binds.Genes in which transcription is prevented are said to be __________.The interaction of small molecules, such as tryptophan, with __________ DNA-binding proteins, such as the tryptophan repressor, regulates bacterial genes.Genes that are being __________ expressed are being transcribed all the time.
(Essay)
4.8/5  (34)
(34)
You are interested in the regulation of gene Q.Proteins G, H, and J are proteins that are important for regulating gene Q, and bind to its promoter region in a sequence-specific fashion.Proteins G and H both bind to site "A" but cannot bind to site "A" at the same time.Protein J binds to site "B" on the promoter.The promoter region is diagrammed in Figure 8-20.  Figure 8-20
You develop a cell-free transcriptional system to study the effects of proteins G, H, and J on the transcription of gene Q.Using this system, you can examine the effects of adding these proteins to the transcriptional system in equal amounts and measuring how much gene Q is produced.When you add these proteins to the system, you get the results shown in Table 8-20.
number G H J made? 1 - - - no 2 + - - no 3 - + - yes 4 - - + yes 5 + + - no 6 + - + yes 7 - + + yes 8 + + + yes
Table 8-20
Which proteins are likely to act as gene activators?
Figure 8-20
You develop a cell-free transcriptional system to study the effects of proteins G, H, and J on the transcription of gene Q.Using this system, you can examine the effects of adding these proteins to the transcriptional system in equal amounts and measuring how much gene Q is produced.When you add these proteins to the system, you get the results shown in Table 8-20.
number G H J made? 1 - - - no 2 + - - no 3 - + - yes 4 - - + yes 5 + + - no 6 + - + yes 7 - + + yes 8 + + + yes
Table 8-20
Which proteins are likely to act as gene activators?
(Multiple Choice)
4.8/5  (37)
(37)
Enhancers can act over long stretches of DNA, but are specific about which genes they affect.How do eukaryotic cells prevent these transcription regulators from looping in the wrong direction and inappropriately turning on the transcription of a neighboring gene?
(Multiple Choice)
4.8/5  (39)
(39)
From the sequencing of the human genome, we believe that there are approximately 21,000 protein-coding genes in the genome, of which 1500-3000 are transcription factors.If every gene has a tissue-specific and signal-dependent transcription pattern, how can such a small number of transcriptional regulatory proteins generate a much larger set of transcriptional patterns?
(Essay)
4.7/5  (40)
(40)
In principle, a eukaryotic cell can regulate gene expression at any step in the pathway from DNA to the active protein.Place the types of control listed below at the appropriate places on the diagram in Figure 8-67. 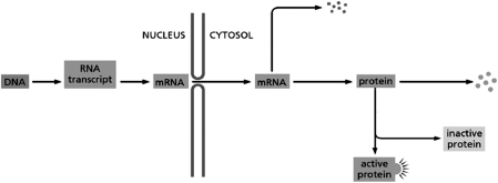 Figure 8-67
A.translation control
B.transcriptional control
C.RNA splicing
D.RNA degradation
E.protein degradation
Figure 8-67
A.translation control
B.transcriptional control
C.RNA splicing
D.RNA degradation
E.protein degradation
(Essay)
4.8/5  (36)
(36)
For each of the following sentences, fill in the blanks with the best word or phrase selected from the list below.Not all words or phrases will be used; use each word or phrase only once.
combinatorial feedback phosphorylation deacetylation histone pluripotency differential leucine zipper proliferation epigenetic memory receptor expression methylation unwinding The transmission of information important for gene regulation from parent to daughter cell, without altering the actual nucleotide sequence, is called __________ inheritance.This type of inheritance is seen with the inheritance of the covalent modifications on __________ proteins bound to DNA; these modifications are important for reestablishing the pattern of chromatin structure found on the parent chromosome.Another way to inherit chromatin structure involves DNA __________, a covalent modification that occurs on cytosine bases that typically turns off the transcription of a gene.Gene transcription patterns can also be transmitted across generations through positive __________ loops that can involve a transcription regulator activating its own transcription in addition to other genes.These mechanisms all allow for cell __________, a property involving the maintenance of gene expression patterns important for cell identity.
(Essay)
4.8/5  (38)
(38)
The human genome encodes about 21,000 protein-coding genes.Approximately how many such genes does the typical differentiated human cell express at any one time?
(Multiple Choice)
4.9/5  (27)
(27)
Which of the following is NOT a general mechanism that cells use to maintain stable patterns of gene expression as cells divide?
(Multiple Choice)
4.8/5  (31)
(31)
Showing 41 - 60 of 68
Filters
- Essay(0)
- Multiple Choice(0)
- Short Answer(0)
- True False(0)
- Matching(0)