Exam 10: Analyzing the Structure and Function of Genes
Exam 1: Cells: The Fundamental Units of Life64 Questions
Exam 2: Chemical Components of Cells74 Questions
Exam 3: Energy, Catalysis, and Biosynthesis73 Questions
Exam 4: Protein Structure and Function71 Questions
Exam 5: DNA and Chromosomes69 Questions
Exam 6: DNA Replication and Repair61 Questions
Exam 7: From DNA to Protein62 Questions
Exam 8: Control of Gene Expression68 Questions
Exam 9: How Genes and Genomes Evolve60 Questions
Exam 10: Analyzing the Structure and Function of Genes59 Questions
Exam 11: Membrane Structure57 Questions
Exam 12: Transport Across Cell Membranes67 Questions
Exam 13: How Cells Obtain Energy From Food71 Questions
Exam 14: Energy Generation in Mitochondria and Chloroplasts72 Questions
Exam 15: Intracellular Compartments and Protein Transport55 Questions
Exam 16: Cell Signaling60 Questions
Exam 17: Cytoskeleton59 Questions
Exam 18: The Cell-Division Cycle67 Questions
Exam 19: Sexual Reproduction and the Power of Genetics61 Questions
Exam 20: Cell Communities: Tissues, Stem Cells, and Cancer57 Questions
Select questions type
You create a recombinant DNA molecule that fuses the coding sequence of green fluorescent protein to the regulatory DNA sequences that control the expression of your favorite genes.Which of the following pieces of information can you NOT gain by examining the expression of this reporter gene?
(Multiple Choice)
4.9/5  (44)
(44)
Your friend works at the Centers for Disease Control and Prevention and has discovered a brand-new virus that has recently been introduced into the human population.She has just developed a new assay that allows her to detect the virus by using PCR products made from the blood of infected patients.The assay uses primers in the PCR assay that hybridize to sequences in the viral genome.
Your friend is distraught because of the result she obtained (see Figure 10-28) when she looked at PCR products made using the blood of three patients suffering from the viral disease, using her own blood, and using a leaf from her petunia plant.
You advise your friend not to panic, as you believe she is missing an important control.Which one of the choices listed below is the best control for clarifying the results of her assay? Explain your answer. 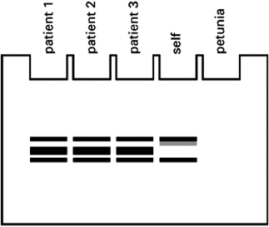 Figure 10-28
Figure 10-28
(Multiple Choice)
4.8/5  (25)
(25)
You are studying a protein, and a small fragment of its sequence is shown below.You have decided that the glutamine in the protein segment has an important role in its function.You decide to change this glutamine to a lysine by changing only one base.Given the partial mRNA sequence that codes for this stretch of protein below, write the new sequence for this stretch of RNA if you were to make this change.Be sure to label the 5′ and 3′ ends.
U C A G UUU Phe (F) UCU Ser (S) UAU Tyr (Y) UGU Cys (C) UUC - UCC - UAC - UGC - U UUA Leu (L) UCA -- UAA Stop UGA Stop UUG - UCG - UAG Stop UGG Trp (W) CUU Leu (L) CCU Pro (P) CAU His (H) CGU Arg (R) CUC - CCC - CAC - CGC - C CUA - CCA - CAA Gin (Q) CGA - CUG - CCG - CAG - CGG - AUU Ile (I) ACU Thr (T) AAU Asn (N) AGU Ser (S) AUC - ACC - AAC - AGC - A AUA - ACA - AAA Lys (K) AGA Arg (R) AUG Met (M) ACG - AAG - AGG - GUU Val (V) GCU Ala (A) GAU Asp (D) GGU Gly (G) GUC - GCC - GAC - GGC - G GUA - GCA - GAA Glu (E) GGA - GUG - GCG - GAG - GGG -
Table 10-56
F D P Q G S H
5′-UUCGACCCGCAGGGCAGCCAC-3′
(Essay)
4.7/5  (41)
(41)
You have a linear piece of DNA that can be cut by the restriction nucleases HindIII and EcoRI, as diagrammed in Figure 10-7.  Figure 10-7
If you were to cut this linear DNA with HindIII, what type of DNA fragments do you predict you would obtain?
Figure 10-7
If you were to cut this linear DNA with HindIII, what type of DNA fragments do you predict you would obtain?
(Multiple Choice)
4.9/5  (39)
(39)
You have a piece of circular DNA that can be cut by the restriction nucleases XhoI and SmaI, as indicated in Figure 10-5. 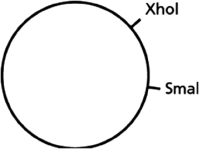 Figure 10-5
If you were to cut this circular piece of DNA with both XhoI and SmaI, how many fragments of DNA would you end up with?
Figure 10-5
If you were to cut this circular piece of DNA with both XhoI and SmaI, how many fragments of DNA would you end up with?
(Multiple Choice)
4.9/5  (35)
(35)
You want to amplify the DNA between the two stretches of sequence shown in Figure 10-58.Of the listed primers, choose the pair that will allow you to amplify the DNA by PCR. 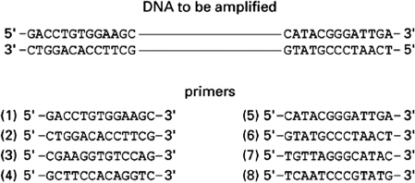 Figure 10-58
Figure 10-58
(Essay)
4.8/5  (29)
(29)
You have been asked to consult for a biotech company that is seeking to understand why some fungi can live in very extreme environments, such as the high temperatures inside naturally occurring hot springs.The company has isolated two different fungal species, F.cattoriae and W.gravinius, both of which can grow at temperatures exceeding 95°C.The company has determined the following things about these fungal species: Property F. cattoriae W. gravinius Genome size 1 3 Repetitive DNA? 20\% of genome contains large stretches of CG repeats <0.1\% of genome
By sequencing and examining their genomes, the biotech company hopes to understand why these species can live in extreme environments.However, the company only has the resources to sequence one genome, and would like your input as to which species should be sequenced and whether you believe a shotgun strategy will work in this case.(Be sure to explain your answer.)
(Essay)
4.7/5  (40)
(40)
Which of the following statements about genomic DNA libraries is FALSE?
(Multiple Choice)
4.9/5  (48)
(48)
Which of the following statements about DNA libraries is TRUE?
(Multiple Choice)
4.8/5  (30)
(30)
You have the amino acid sequence of a protein and wish to design PCR primers that will allow you to amplify this gene by PCR from genomic DNA.Using this protein sequence, you deduce a particular DNA sequence that can encode this protein.Why is it unwise to use only this DNA sequence you have deduced to design PCR primers for isolating the gene encoding your protein of interest from genomic DNA?
(Essay)
4.9/5  (35)
(35)
You want to design a set of PCR primers to amplify a portion of a gene from a cDNA library.Which of the following concerns about PCR primer design is the most legitimate?
(Multiple Choice)
4.9/5  (43)
(43)
DNA ligase is an enzyme used when making recombinant DNA molecules in the lab.In what normal cellular process is DNA ligase involved?
(Multiple Choice)
4.9/5  (33)
(33)
You have a piece of circular DNA that can be cut by the restriction nucleases EcoRI, HindIII, and NotI, as indicated in Figure 10-6. 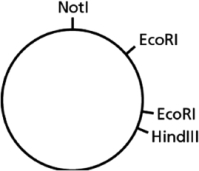 Figure 10-6
Which of the following statements is FALSE?
Figure 10-6
Which of the following statements is FALSE?
(Multiple Choice)
4.9/5  (37)
(37)
Why are dideoxyribonucleoside triphosphates used during DNA sequencing?
(Multiple Choice)
4.9/5  (30)
(30)
PCR involves a heating step, followed by a cooling step, and then DNA synthesis.What is the primary reason for why this cooling step is necessary?
(Multiple Choice)
4.9/5  (40)
(40)
Which of the following statements about RNA interference (or RNAi) is FALSE?
(Multiple Choice)
4.7/5  (23)
(23)
For each of the following sentences, fill in the blanks with the best word or phrase selected from the list below.Not all words or phrases will be used; use each word or phrase only once.
DNA sequencing polymerase chain reaction DNA ligase recombinant DNA endonucleases restriction enzymes gel electrophoresis ribonucleases
Nucleases that cut DNA only at specific short sequences are known as __________.DNA composed of sequences from different sources is known as __________.__________ can be used to separate DNA fragments of different sizes.Millions of copies of a DNA sequence can be made entirely in vitro by the __________ technique.
(Essay)
4.8/5  (45)
(45)
With fully automated Sanger sequencing, all four chain-terminating ddNTPs are added into a single reaction and loaded onto a capillary gel.The sequence can be determined even though all ddNTPs are mixed together because
(Multiple Choice)
4.9/5  (36)
(36)
Figure 10-45A depicts the restriction map of one segment of the human genome for four restriction nucleases W, X, Y, and Z.Figure 10-45B depicts the restriction maps of four individual BAC clones that contain segments of human DNA from the region depicted in Figure 10-45A. (A)
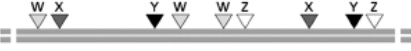 (B)
(B)
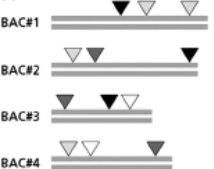 Figure 10-45
From this information, how would you order these BAC clones, from left to right?
Figure 10-45
From this information, how would you order these BAC clones, from left to right?
(Multiple Choice)
4.9/5  (34)
(34)
Showing 21 - 40 of 59
Filters
- Essay(0)
- Multiple Choice(0)
- Short Answer(0)
- True False(0)
- Matching(0)