Exam 13: Amplification Techniques
Exam 1: What Is Immunology45 Questions
Exam 2: Overview of the Immune System43 Questions
Exam 3: Immunoglobulins45 Questions
Exam 4: Infection and Immunity: Hostparasite Relationships45 Questions
Exam 5: The Complement System45 Questions
Exam 6: Hypersensitivity45 Questions
Exam 7: Immunologic Detection of Infectious Diseases45 Questions
Exam 8: Autoimmune Diseases and Immunodeficiency Disorders45 Questions
Exam 9: Transplant Immunology45 Questions
Exam 10: Tumor Immunology45 Questions
Exam 11: Clinical Applications of Flow Cytometry45 Questions
Exam 12: Hybridization Techniques45 Questions
Exam 13: Amplification Techniques45 Questions
Exam 14: Molecular Applications in Cytogenetics45 Questions
Exam 15: Histocompatibility Laboratory Techniques and Applications45 Questions
Select questions type
The three basic steps of polymerase chain reaction (PCR)are denaturation, primer annealing, and primer extension. Which step requires DNA polymerase?
(Multiple Choice)
5.0/5  (42)
(42)
Since Taq polymerase cannot function without free magnesium ions (Mg++)and many chemical species can bind Mg++, a gross excess of Mg++ must always be added to any PCR reaction mixture.
(True/False)
4.8/5  (47)
(47)
mRNA preparations that have been treated with diethylpyrocarbonate (DEPC)can usually stored several weeks in the freezer.
(True/False)
4.9/5  (30)
(30)
Explain how reverse transcriptase PCR can be used to accurately quantify RNA in different samples.
(Essay)
4.8/5  (48)
(48)
RNA- dependent DNA polymerase is the proper name for the enzyme ________.
(Short Answer)
4.9/5  (33)
(33)
What advantage does DNA polymerase from Thermus aquaticus (Taq polymerase)have over DNA polymerase from E. coli?
(Multiple Choice)
4.9/5  (32)
(32)
When making mRNA preparations for reverse transcriptase PCR, it is most important to eliminate contamination from
(Multiple Choice)
4.9/5  (41)
(41)
Even a small amount of contaminating nucleic acid can cause a false positive PCR result.
(True/False)
4.8/5  (36)
(36)
How can one quantify the amount of nucleic acid that is present in a sample and determine its purity?
(Essay)
4.8/5  (30)
(30)
Reverse transcriptase PCR can only be done on pure preparations of messenger RNA as ribosomal RNA and transfer RNA interfere significantly.
(True/False)
4.8/5  (37)
(37)
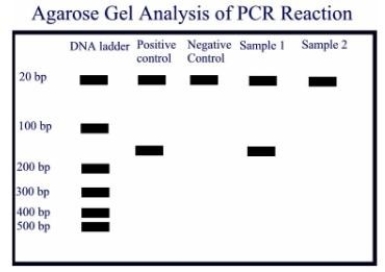 -A PCR amplification was done on a positive control containing the template DNA, a negative control without the template DNA and two samples. Oligonucleotide primers of 21 base pairs were used, and the expected amplicon product is about 160 base pairs. Observe the agarose gel electrophoresis done on the amplicons, and give the most appropriate interpretations.
11eae1fd_ee0f_a6d4_ad0d_15a6cfbbac73_TB4770_00
-A PCR amplification was done on a positive control containing the template DNA, a negative control without the template DNA and two samples. Oligonucleotide primers of 21 base pairs were used, and the expected amplicon product is about 160 base pairs. Observe the agarose gel electrophoresis done on the amplicons, and give the most appropriate interpretations.
11eae1fd_ee0f_a6d4_ad0d_15a6cfbbac73_TB4770_00
(Multiple Choice)
4.8/5  (38)
(38)
A standard reverse transcriptase PCR assay uses ________(how many?)primer(s)to convert mRNA to cDNA.
(Short Answer)
4.9/5  (47)
(47)
Review the protocol herein and identify its name. Two labeled probes attached to target DNA. DNA polymerase fills in the nucleotides between the p and DNA ligase joins the two pieces. The product is denatured into single strands, and the process i repeated. Amplified target DNA is detected via the labels on the probes.
(Multiple Choice)
4.8/5  (36)
(36)
Taq polymerase can add nucleotide bases to a DNA strand at a rate of about
(Multiple Choice)
4.8/5  (38)
(38)
Which amplification technique listed is best to use if one wishes to characterize the 5' end of an mRNA transcript?
(Multiple Choice)
4.9/5  (33)
(33)
How does one assure that all PCR amplicons are double- stranded?
(Multiple Choice)
4.8/5  (39)
(39)
Showing 21 - 40 of 45
Filters
- Essay(0)
- Multiple Choice(0)
- Short Answer(0)
- True False(0)
- Matching(0)