Exam 4: Mapping Eukaryote Chromosomes by Recombination
Exam 1: The Genetics Revolution22 Questions
Exam 2: Single-Gene Inheritance51 Questions
Exam 3: Independent Assortment of Genes55 Questions
Exam 4: Mapping Eukaryote Chromosomes by Recombination64 Questions
Exam 5: The Genetics of Bacteria and Their Viruses44 Questions
Exam 6: Gene Interaction47 Questions
Exam 7: Dna: Structure and Replication50 Questions
Exam 8: Rna: Transcription and Processing53 Questions
Exam 9: Proteins and Their Synthesis53 Questions
Exam 10: Gene Isolation and Manipulation55 Questions
Exam 11: Regulation of Gene Expression in Bacteria and Their Viruses56 Questions
Exam 12: Regulation of Gene Expression in Eukaryotes46 Questions
Exam 13: The Genetic Control of Development36 Questions
Exam 14: Genomes and Genomics26 Questions
Exam 15: The Dynamic Genome: Transposable Elements32 Questions
Exam 16: Mutation, Repair, and Recombination53 Questions
Exam 17: Large-Scale Chromosomal Changes50 Questions
Exam 18: Population Genetics48 Questions
Exam 19: The Inheritance of Complex Traits36 Questions
Exam 20: Evolution of Genes and Traits54 Questions
Select questions type
An experiment was done to determine the linkage relationship of three genes (a,b,and c)in Drosophila melanogaster.Homozygous females phenotypically a,c were crossed with homozygous males phenotypically b.The F1 females were all wild type in appearance,and the F1 males were all a,c.The F1 females and males were crossed to give the following F2 phenotypes and numbers.
Males Females a+c 242 310 +b+ 238 0 ab+ 157 0 ++c 163 190 abc 59 0 +++ 61 295 a++ 38 205 +bc 42 0 Total 1,000 1,000
a)Give the genotypes of the parents.
b)Give the genotypes of the F1 flies,and show the genotypic arrangement in the female F1.
c)Draw a genetic map showing the order of the genes and the distances between them.
d)Calculate the interference (only if necessary;if not,say "I = 0").
Free
(Essay)
4.9/5  (29)
(29)
Correct Answer:
The data clearly show sex linkage due to disproportionate representation of phenotypic classes between the sexes.
a) 
b)  11eee395_9261_0cf6_9538_ef5bfdb5100b_TB3928_11
11eee395_9261_0cf6_9538_ef5bfdb5100b_TB3928_11
Loci order must be because the most rare F2 classes of 38 and 42 flies each (due to D-CO)are a++ and +bc.
c)a-b: Look for recombinants (157 + 59)and + + (163 + 61)Total = 440
RF = 440/1000 = 44%
a-c: Look for recombinants + (157 + 38)and + (163 + 42)Total = 400
RF = 400/1000 = 0.40 or 40%
b-c: Look for recombinants (59 + 42)and + + (61 + 38)Total = 200
RF = 200/1000 = 0.20 or 20%
d)Expected D-CO = 0.4 * 0.2 *1000 = 80
Observed D-CO = 80;therefore,Interference (I)= 0.The correct answer is
"I = 0."
Neurospora loci d and f are located on the same chromosome.The d locus always segregates at MI,while the e locus segregates at MII at a frequency of 10%.Assuming complete interference,the expected frequency of parental ditypes in the progeny of a cross d+e+ d e is:
Free
(Multiple Choice)
4.8/5  (31)
(31)
Correct Answer:
E
A dihybrid in cis is crossed to a dihybrid in trans (the two genes of interest,A and B,are linked).What proportion of the progeny is expected to be homozygous recessive with respect to the two genes of interest?
Free
(Multiple Choice)
4.9/5  (27)
(27)
Correct Answer:
A
The molecular process which results in the formation of recombinant chromosomes is initiated by:
(Multiple Choice)
4.7/5  (45)
(45)
In Drosophila,the genes crossveinless-c and Stubble are linked,about 7 map units apart on chromosome 3.cv-c is a recessive mutant allele of crossveinless-c (cv-c+ is wild type),while Sb is a dominant mutant allele of Stubble (Sb+ is wild type). A dihybrid female Drosophila with genotype cv-c Sb+/cv-c+Sb is testcrossed.The proportion of phenotypically wild-type individuals in the progeny of the testcross will be:
(Multiple Choice)
4.7/5  (48)
(48)
Out of 800 progeny of a three-point testcross,there were 16 double crossover recombinants,whereas 80 had been expected on the basis of no interference.The interference is:
(Multiple Choice)
4.9/5  (39)
(39)
In unordered tetrad analyses of two linked loci,a nonparental ditype is an indication of:
(Multiple Choice)
4.8/5  (29)
(29)
In one short chromosome arm the average frequency of crossovers per meiosis was measured to be 0.5.What proportion of meioses will have no crossovers at all in this region?
(Multiple Choice)
4.9/5  (29)
(29)
In the cross mt+al+ mt al,the mt locus shows an MII frequency of 12%,the al locus an MII frequency of 10%.Most asci were parental ditypes.The order of loci is:
(Multiple Choice)
4.9/5  (35)
(35)
Draw a linkage map or maps consistent with the following recombination frequencies:
a)A-B 30%,A-C 50%,B-C 50%,B-D 50%,A-D 50%,C-D 15%
b)A-B 30%,A-C 50%,B-C 20%,B-D 35%,A-D 50%,C-D 15%
(Essay)
4.9/5  (39)
(39)
The maize genes sh and bz are linked,40 map units apart.If a plant sh+bz/sh bz+ is selfed,what proportion of the progeny will be sh bz/sh bz?
(Multiple Choice)
4.7/5  (31)
(31)
In an unordered ascus analysis of the Neurospora loci cot and fl,there were 42 tetratypes and 8 nonparental ditypes out of a total of 290 asci.These loci are linked at a distance of:
(Multiple Choice)
4.8/5  (41)
(41)
A dihybrid AaBb female Drosophila is testcrossed with an aabb male.The following offspring genotypes were obtained.
AaBb 99
Aabb 83
AaBb 86
Aabb 92
A 2 test was performed to determine whether the data supported the original hypothesis that the genes are unlinked.How many degrees of freedom should be used in this analysis?
(Multiple Choice)
4.8/5  (37)
(37)
In a linear tetrad analysis,the second division segregation (MII)frequency of the cyh locus is 16%.The map distance from this locus to its centromere is:
(Multiple Choice)
4.9/5  (31)
(31)
A sweet pea plant of genotype A/A . B/B is crossed to one that is a/a . b/b to produce a dihybrid F1.One cell of one F1 individual (genotype A/a . B/b)goes through meiosis and produces four gametes of the following genotypes: A . b,A . b,a .B and a .B.What can be concluded regarding the linkage relationship between the two genes?
(Multiple Choice)
4.8/5  (37)
(37)
The maximum second division segregation frequency normally possible is:
(Multiple Choice)
4.9/5  (39)
(39)
A plant of genotype C/C ;d/d is crossed to c/c ;D/D,and the F1 is testcrossed.If the genes in question are unlinked,the percentage of double homozygous recessive individuals in the offspring of the testcross will be:
(Multiple Choice)
5.0/5  (42)
(42)
Below is a pedigree for a very rare,late-onset genetic condition.Below the pedigree are represented the RFLP banding patterns obtained for each individual in the family.The RFLP locus used is about 2 cM away from the gene responsible for the disease.Note that darker/thicker bands indicate homozygosity for that particular morph of the RFLP. 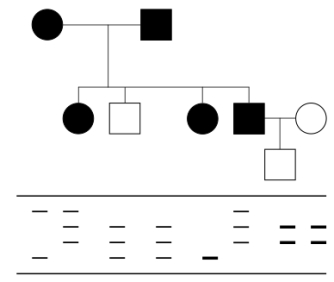 What is the probability that the child in generation III will develop the disease?
What is the probability that the child in generation III will develop the disease?
(Multiple Choice)
4.9/5  (34)
(34)
In a haploid organism,the loci leu and arg are linked,30 map units apart.In a cross of leu+arg * leu arg+,what proportion of progeny will be leu arg?
(Multiple Choice)
4.8/5  (40)
(40)
Showing 1 - 20 of 64
Filters
- Essay(0)
- Multiple Choice(0)
- Short Answer(0)
- True False(0)
- Matching(0)