Exam 10: Gene Isolation and Manipulation
Exam 1: The Genetics Revolution22 Questions
Exam 2: Single-Gene Inheritance51 Questions
Exam 3: Independent Assortment of Genes55 Questions
Exam 4: Mapping Eukaryote Chromosomes by Recombination64 Questions
Exam 5: The Genetics of Bacteria and Their Viruses44 Questions
Exam 6: Gene Interaction47 Questions
Exam 7: Dna: Structure and Replication50 Questions
Exam 8: Rna: Transcription and Processing53 Questions
Exam 9: Proteins and Their Synthesis53 Questions
Exam 10: Gene Isolation and Manipulation55 Questions
Exam 11: Regulation of Gene Expression in Bacteria and Their Viruses56 Questions
Exam 12: Regulation of Gene Expression in Eukaryotes46 Questions
Exam 13: The Genetic Control of Development36 Questions
Exam 14: Genomes and Genomics26 Questions
Exam 15: The Dynamic Genome: Transposable Elements32 Questions
Exam 16: Mutation, Repair, and Recombination53 Questions
Exam 17: Large-Scale Chromosomal Changes50 Questions
Exam 18: Population Genetics48 Questions
Exam 19: The Inheritance of Complex Traits36 Questions
Exam 20: Evolution of Genes and Traits54 Questions
Select questions type
Bacterial cells have a variety of restriction enzymes that can degrade the DNA of an attacking virus.How do these bacterial cells protect their own DNA from being degraded by the very restriction enzymes that are meant to help them?
(Essay)
4.8/5  (37)
(37)
Which of the following organisms can be made transgenic by injection of DNA into the gonad,generating extrachromosomal arrays of the transgene in some egg cells?
(Multiple Choice)
4.9/5  (41)
(41)
The drawing below depicts a restriction map of a segment of a human chromosome.The cut sites shown (1 through 6)are known to be polymorphic and can be present or absent depending on the genetic makeup of a specific individual's chromosomes.E identifies EcoRI sites,and P identifies Pst1 sites.The line below the map identifies the region of homology between the DNA and a probe used in Southern blot analyses to characterize this region. 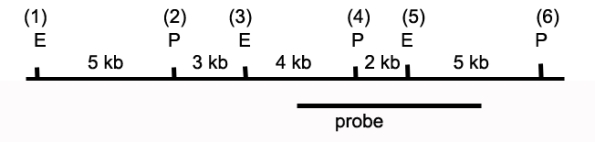 a)Individual 1 has restriction sites 1 through 6 (all)on both copies of this chromosome.If DNA from this individual was digested with Pst I,what are the sizes of the fragments that would be detected via Southern blot using the identified probe?
b)DNA from Individual 1 is digested with both EcoRI and PstI and then analyzed by Southern blot.What bands are revealed?
c)Individual 2 is homozygous for a polymorphism that eliminates site 3.If DNA from this individual was digested with both PstI and EcoRI,what are the sizes of the fragments that would be detected via Southern blot?
d)Individual 3 has not previously been characterized over this chromosomal locus.The research team digests the DNA with PstI and EcoRI and generates the following banding pattern.What is the likely genotype of this individual?
a)Individual 1 has restriction sites 1 through 6 (all)on both copies of this chromosome.If DNA from this individual was digested with Pst I,what are the sizes of the fragments that would be detected via Southern blot using the identified probe?
b)DNA from Individual 1 is digested with both EcoRI and PstI and then analyzed by Southern blot.What bands are revealed?
c)Individual 2 is homozygous for a polymorphism that eliminates site 3.If DNA from this individual was digested with both PstI and EcoRI,what are the sizes of the fragments that would be detected via Southern blot?
d)Individual 3 has not previously been characterized over this chromosomal locus.The research team digests the DNA with PstI and EcoRI and generates the following banding pattern.What is the likely genotype of this individual? 
(Essay)
4.9/5  (35)
(35)
Which of the following about the Sanger sequencing method is/are TRUE?
(Multiple Choice)
4.8/5  (34)
(34)
Restriction endonucleases BamH1,NheI,XbaI,and EcoRI make sequence-specific cuts in DNA at the following sequences:
BamHI: 5'-GGATCC-3'
NheI: 5'-GCTAGC-3'
XbaI: 5'-TCTAGA-3'
EcoRI: 5'-GAATTC-3'
All these enzymes break a phosphodiester bond between the first and second nucleotide (counting from the 5' end),leaving a 5' P and a 3' OH.
Could ligase covalently join two ends produced by the following enzymes? If so,would ligation regenerate one of the above restriction sites?
(1)two BamH1 ends?
(2)two XbaI ends?
(3)a BamH1 and an EcoRI end?
(4)an XbaI and an NheI end?
(5)a BamH1 and an NheI end?
(Essay)
4.9/5  (27)
(27)
A linear molecule of DNA 9 kb in length is isolated and mapped using restriction endonucleases EcoRI and HindIII.The fragments generated by the single and double digests are shown below.The numbers refer to the length of fragments in kilobases.
EcoRI: 7 kb,2 kb
HindIII: 5 kb,4 kb
EcoRI + HindIII: 4 kb,3 kb,2 kb
SalI: 8.5 kb,0.5 kb
EcoRI + SalI: 7 kb,1.5 kb,0.5 kb
Construct a restriction map of this linear fragment of DNA.
(Essay)
4.8/5  (28)
(28)
A plasmid vector has a gene for erythromycin resistance (EryR)and a gene for ampicillin resistance (AmpR).The Amp gene is cut with restriction enzyme,and donor DNA treated with the same enzyme is added.What genotype of cells needs to be selected to show evidence of transformation?
(Multiple Choice)
4.9/5  (48)
(48)
In the Sanger sequencing method,the use of dideoxy adenosine triphosphate stops nucleotide polymerization opposite:
(Multiple Choice)
4.8/5  (39)
(39)
The autoradiogram shown was generated by the dideoxy sequencing method.Define the DNA sequence of the template strand that was used in this sequencing reaction,noting the 5′ and 3′ ends of this template strand. 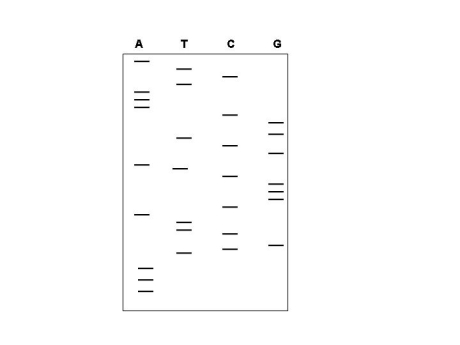
(Essay)
4.7/5  (39)
(39)
The intermediate temperature cycle (72°C)in the polymerase chain reaction enables:
(Multiple Choice)
4.7/5  (41)
(41)
A chimeric plasmid contains vector DNA ligated to a fragment of chromosomal mouse DNA that is suspected of containing the obese gene.As a first step,a restriction map of the DNA clone was constructed.The DNA was digested with two restriction enzymes,BopI and ZapII,with the following results:
 a)From these data,answer the following questions.
(1)How large is the entire DNA clone?
(2)Is the clone linear or circular?
(3)Draw a restriction map consistent with these data.Include all BopI and ZapII sites and the physical distances between them.
b)To better identify the location of the mouse chromosomal DNA on this chimeric plasmid,mouse genomic DNA was digested with ZapII,run on a gel,and analyzed using the Southern blot technique.The Southern blot was probed with radiolabeled Zap II DNA fragments from the plasmid DNA.Three lanes each containing the same amount of Zap II-digested chromosomal DNA were each probed with a different fragment from the plasmid.Autoradiograms showed the following: What is the maximum amount of mouse DNA present in the clone? Why didn't the 2.0-kb ZapII fragment hybridize to the mouse DNA?
a)From these data,answer the following questions.
(1)How large is the entire DNA clone?
(2)Is the clone linear or circular?
(3)Draw a restriction map consistent with these data.Include all BopI and ZapII sites and the physical distances between them.
b)To better identify the location of the mouse chromosomal DNA on this chimeric plasmid,mouse genomic DNA was digested with ZapII,run on a gel,and analyzed using the Southern blot technique.The Southern blot was probed with radiolabeled Zap II DNA fragments from the plasmid DNA.Three lanes each containing the same amount of Zap II-digested chromosomal DNA were each probed with a different fragment from the plasmid.Autoradiograms showed the following: What is the maximum amount of mouse DNA present in the clone? Why didn't the 2.0-kb ZapII fragment hybridize to the mouse DNA? 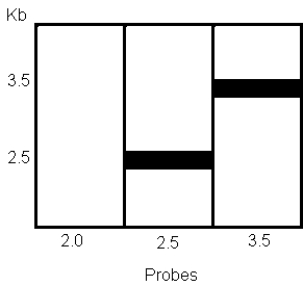
(Essay)
4.9/5  (44)
(44)
The cDNA for a eukaryotic gene B is 900 nucleotide pairs long.A cDNA clone is used to isolate a genomic clone of gene B,and the gene is sequenced.From start to stop codon,the gene is found to be 1800 nucleotide pairs long.The most probable reason for the discrepancy is that:
(Multiple Choice)
4.9/5  (39)
(39)
Recombinant DNA techniques typically require the action of:
(Multiple Choice)
4.9/5  (38)
(38)
In Southern and Northern blotting,the probe being used to analyze DNA or RNA identifies the target sequence via:
(Multiple Choice)
4.7/5  (32)
(32)
A linear DNA molecule has n target sites for restriction enzyme EcoRI.How many fragments will be produced after complete digestion?
(Multiple Choice)
4.9/5  (40)
(40)
Human insulin can be made purer,and at a lower cost,using recombinant DNA technology.What sort of DNA clone might be ligated into a bacterial expression plasmid to be used in this strategy?
(Essay)
4.8/5  (30)
(30)
A radioactive probe is generated using the actin gene from yeast.This probe is used in a Southern blot analysis of EcoRI digested genomic DNA from the ciliated protozoan Tetrahymena thermophila.The autoradiogram shows a single-labeled band of 4 kb in size.This means that the Tetrahymena actin gene is:
(Multiple Choice)
4.8/5  (35)
(35)
Showing 21 - 40 of 55
Filters
- Essay(0)
- Multiple Choice(0)
- Short Answer(0)
- True False(0)
- Matching(0)